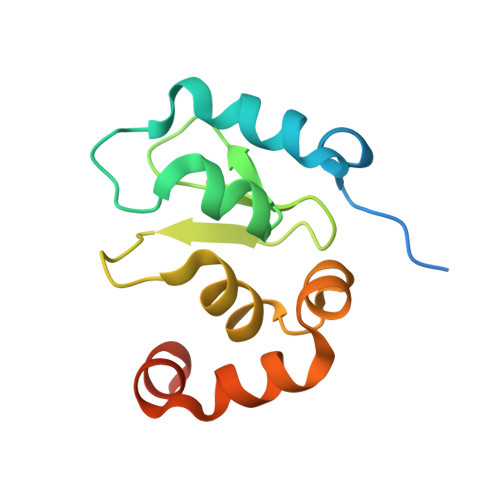
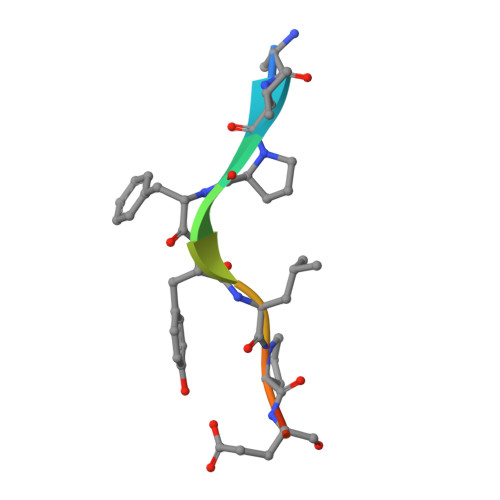
Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Wu, J.W., Cocina, A.E., Chai, J., Hay, B.A., Shi, Y.(2001) Mol Cell 8: 95-104
- PubMed: 11511363
- DOI: https://doi.org/10.1016/s1097-2765(01)00282-9
- Primary Citation of Related Structures:
1JD4, 1JD5, 1JD6 - PubMed Abstract:
The inhibitor of apoptosis protein DIAP1 suppresses apoptosis in Drosophila, with the second BIR domain (BIR2) playing an important role. Three proteins, Hid, Grim, and Reaper, promote apoptosis, in part by binding to DIAP1 through their conserved N-terminal sequences. The crystal structures of DIAP1-BIR2 by itself and in complex with the N-terminal peptides from Hid and Grim reveal that these peptides bind a surface groove on DIAP1, with the first four amino acids mimicking the binding of the Smac tetrapeptide to XIAP. The next 3 residues also contribute to binding through hydrophobic interactions. Interestingly, peptide binding induces the formation of an additional alpha helix in DIAP1. Our study reveals the structural conservation and diversity necessary for the binding of IAPs by the Drosophila Hid/Grim/Reaper and the mammalian Smac proteins.
- Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544, USA.
Organizational Affiliation: