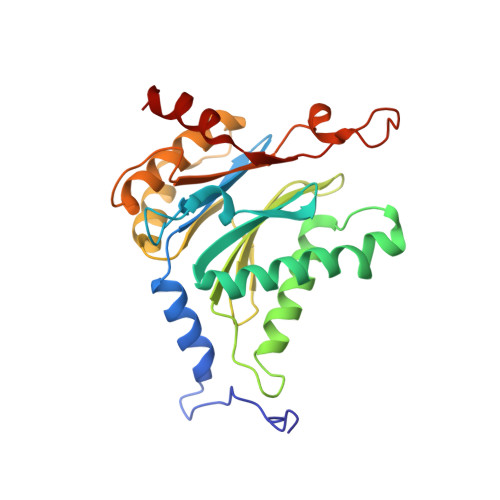
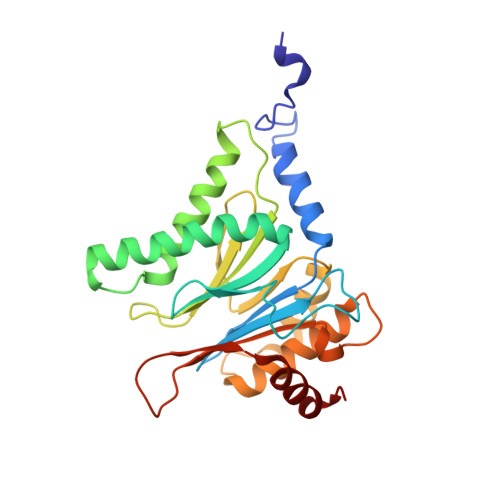
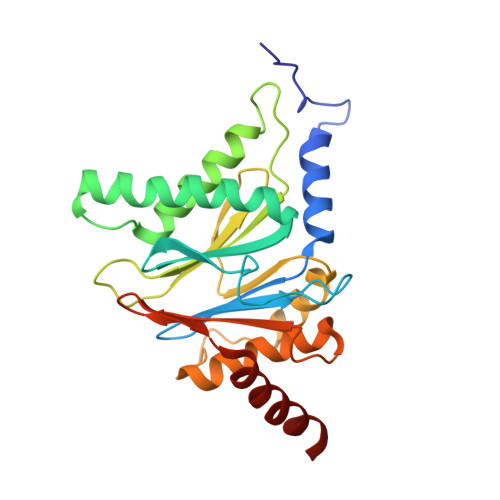
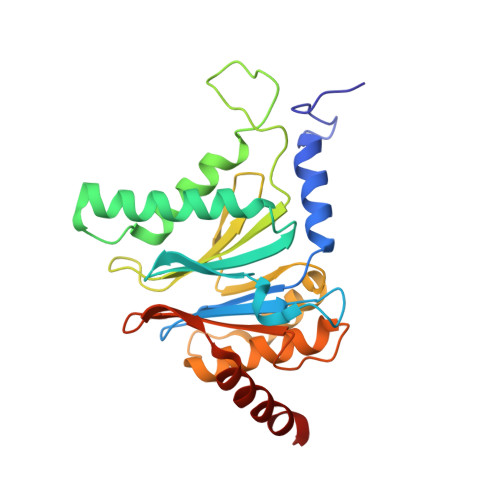
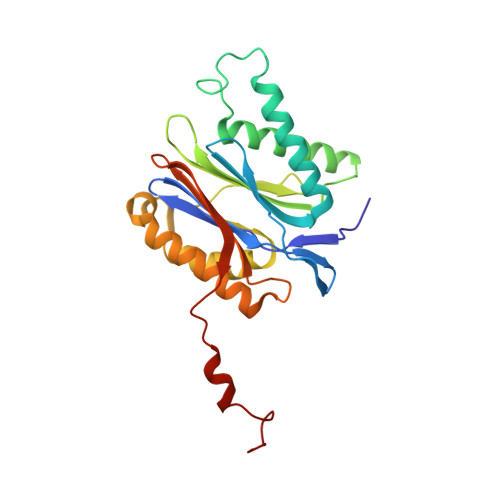
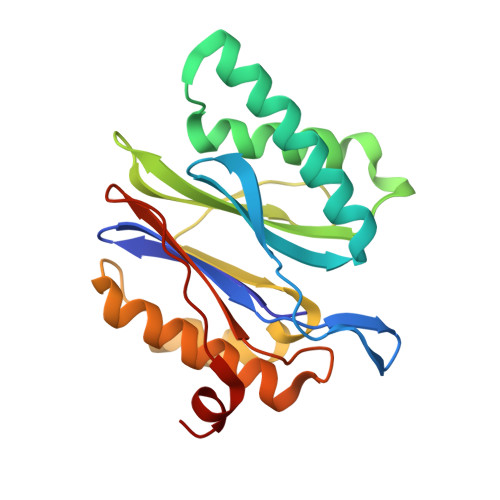
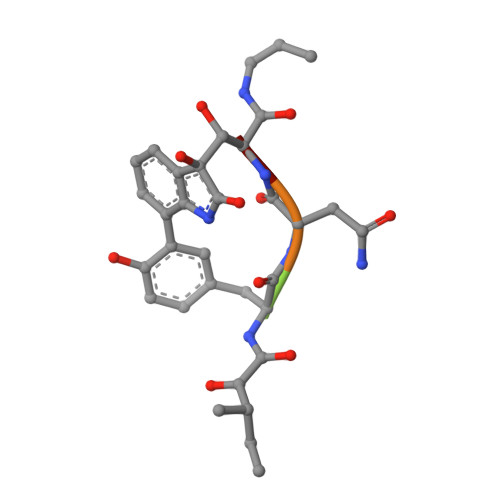
Crystal structure of the 20 S proteasome:TMC-95A complex: a non-covalent proteasome inhibitor.
Groll, M., Koguchi, Y., Huber, R., Kohno, J.(2001) J Mol Biology 311: 543-548
- PubMed: 11493007
- DOI: https://doi.org/10.1006/jmbi.2001.4869
- Primary Citation of Related Structures:
1JD2 - PubMed Abstract:
The 20 S proteasome core particle (CP), a multicatalytic protease, is involved in a variety of biologically important processes, including immune response, cell-cycle control, metabolic adaptation, stress response and cell differentiation. Therefore, selective inhibition of the CP will be one possible way to influence these essential pathways. Recently, a new class of specific proteasome inhibitors, TMC-95s, was investigated and we now present a biochemical and crystallographic characterisation of the yeast proteasome core particle in complex with the natural product TMC-95A. This unusual heterocyclic compound specifically blocks the active sites of CPs non-covalently, without modifying the nucleophilic Thr1 residue. The inhibitor is bound to the CP by specific hydrogen bonds with the main-chain atoms of the protein. Analysis of the crystal structure of the complex has revealed which portions of TMC-95s are essential for binding to the proteasome. This will form the basis for the development of synthetic selective proteasome inhibitors as promising candidates for anti-tumoral or anti-inflammatory drugs.
- Max-Planck-Institut für Biochemie, Martinsried, D-82152, Germany. groll@biochem.mpg.de
Organizational Affiliation: