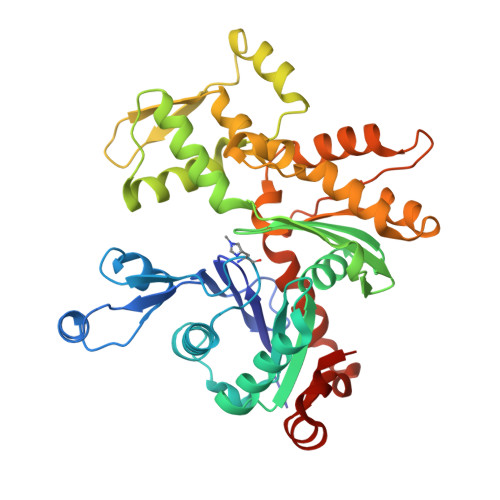
The crystal structure of uncomplexed actin in the ADP state.
Otterbein, L.R., Graceffa, P., Dominguez, R.(2001) Science 293: 708-711
- PubMed: 11474115
- DOI: https://doi.org/10.1126/science.1059700
- Primary Citation of Related Structures:
1J6Z - PubMed Abstract:
The dynamics and polarity of actin filaments are controlled by a conformational change coupled to the hydrolysis of adenosine 5'-triphosphate (ATP) by a mechanism that remains to be elucidated. Actin modified to block polymerization was crystallized in the adenosine 5'-diphosphate (ADP) state, and the structure was solved to 1.54 angstrom resolution. Compared with previous ATP-actin structures from complexes with deoxyribonuclease I, profilin, and gelsolin, monomeric ADP-actin is characterized by a marked conformational change in subdomain 2. The successful crystallization of monomeric actin opens the way to future structure determinations of actin complexes with actin-binding proteins such as myosin.
- Boston Biomedical Research Institute, 64 Grove Street, Watertown, MA 02472, USA.
Organizational Affiliation: