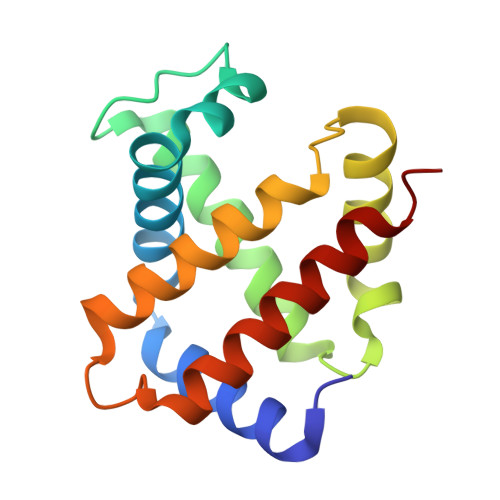
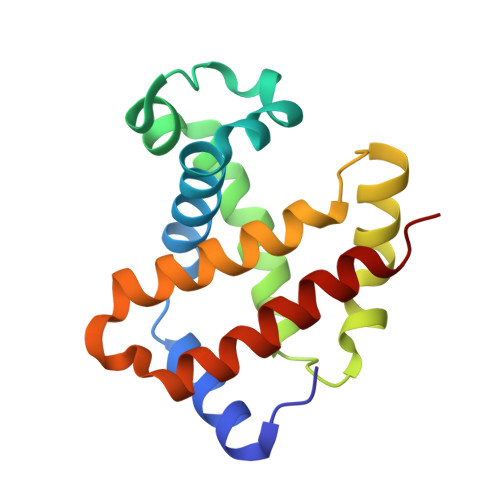
Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Adachi, S., Park, S.-Y., Tame, J.R.H., Shiro, Y., Shibayama, N.(2003) Proc Natl Acad Sci U S A 100: 7039-7044
- PubMed: 12773618
- DOI: https://doi.org/10.1073/pnas.1230629100
- Primary Citation of Related Structures:
1J3Y, 1J3Z, 1J40, 1J41 - PubMed Abstract:
Human Hb, an alpha2beta2 tetrameric oxygen transport protein that switches from a T (tense) to an R (relaxed) quaternary structure during oxygenation, has long served as a model for studying protein allostery in general. Time-resolved spectroscopic measurements after photodissociation of CO-liganded Hb have played a central role in exploring both protein dynamical responses and molecular cooperativity, but the direct visualization and the structural consequences of photodeligation have not yet been reported. Here we present an x-ray study of structural changes induced by photodissociation of half-liganded T-state and fully liganded R-state human Hb at cryogenic temperatures (25-35 K). On photodissociation of CO, structural changes involving the heme and the F-helix are more marked in the alpha subunit than in the beta subunit, and more subtle in the R state than in the T state. Photodeligation causes a significant sliding motion of the T-state beta heme. Our results establish that the structural basis of the low affinity of the T state is radically different between the subunits, because of differences in the packing and chemical tension at the hemes.
- RIKEN Harima Institute/SPring-8, 1-1-1 Kouto, Mikazuki, Sayo, Hyogo 679-5148, Japan. sadachi@spring8.or.jp
Organizational Affiliation: