High Precision NMR Structure and Function of the RING-H2 Finger Domain of EL5, a Rice Protein Whose Expression Is Increased upon Exposure to Pathogen-derived Oligosaccharides
Katoh, S., Hong, C., Tsunoda, Y., Murata, K., Takai, R., Minami, E., Yamazaki, T., Katoh, E.(2003) J Biological Chem 278: 15341-15348
- PubMed: 12588869
- DOI: https://doi.org/10.1074/jbc.M210531200
- Primary Citation of Related Structures:
1IYM - PubMed Abstract:
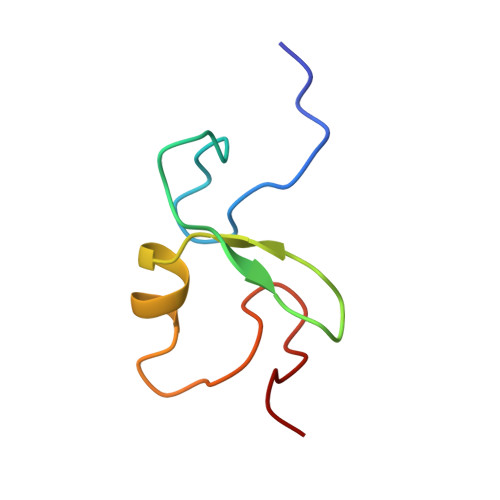
EL5, a RING-H2 finger protein, is rapidly induced by N-acetylchitooligosaccharides in rice cell. We expressed the EL5 RING-H2 finger domain in Escherichia coli and determined its structure in solution by NMR spectroscopy. The EL5 RING-H2 finger domain consists of two-stranded beta-sheets (beta1, Ala(147)-Phe(149); beta2, Gly(156)-His(158)), one alpha-helix (Cys(161)-Leu(166)), and two large N- and C-terminal loops. It is stabilized by two tetrahedrally coordinated zinc ions. This structure is similar to that of other RING finger domains of proteins of known function. From structural analogies, we inferred that the EL5 RING-H2 finger is a binding domain for ubiquitin-conjugating enzyme (E2). The binding site is probably formed by solvent-exposed hydrophobic residues of the N- and C-terminal loops and the alpha-helix. We demonstrated that the fusion protein with EL5-(96-181) and maltose-binding protein (MBP) was polyubiquitinated by incubation with ubiquitin, ubiquitin-activating enzyme (E1), and a rice E2 protein, OsUBC5b. This supported the idea that the EL5 RING finger domain is essential for ubiquitin-ligase activity of EL5. By NMR titration experiments, we identified residues that are critical for the interaction between the EL5 RING-H2 finger and OsUBC5b. We conclude that the RING-H2 finger domain of EL5 is the E2 binding site of EL5.
- Biochemistry Department, National Institute of Agrobiological Sciences, Tsukuba, Ibaraki 305-8602, Japan.
Organizational Affiliation: