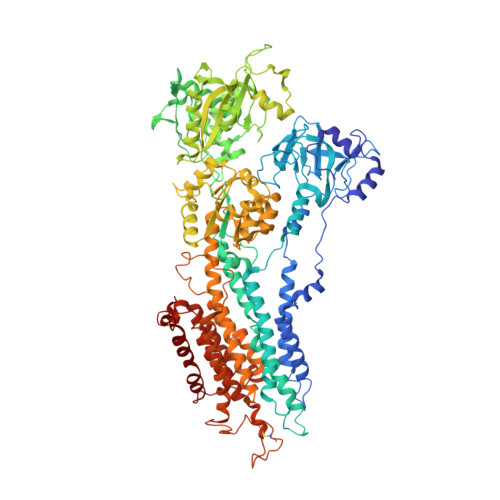
Structural changes in the calcium pump accompanying the dissociation of calcium
Toyoshima, C., Nomura, H.(2002) Nature 418: 605-611
- PubMed: 12167852
- DOI: https://doi.org/10.1038/nature00944
- Primary Citation of Related Structures:
1IWO - PubMed Abstract:
In skeletal muscle, calcium ions are transported (pumped) against a concentration gradient from the cytoplasm into the sarcoplasmic reticulum, an intracellular organelle. This causes muscle cells to relax after cytosolic calcium increases during excitation. The Ca(2+) ATPase that carries out this pumping is a representative P-type ion-transporting ATPase. Here we describe the structure of this ion pump at 3.1 A resolution in a Ca(2+)-free (E2) state, and compare it with that determined previously for the Ca(2+)-bound (E1Ca(2+)) state. The structure of the enzyme stabilized by thapsigargin, a potent inhibitor, shows large conformation differences from that in E1Ca(2+). Three cytoplasmic domains gather to form a single headpiece, and six of the ten transmembrane helices exhibit large-scale rearrangements. These rearrangements ensure the release of calcium ions into the lumen of sarcoplasmic reticulum and, on the cytoplasmic side, create a pathway for entry of new calcium ions.
- Institute of Molecular and Cellular Biosciences, The University of Tokyo, Bunkyo-ku, Tokyo 113-0032, Japan. ct@iam.u-tokyo.ac.jp
Organizational Affiliation: