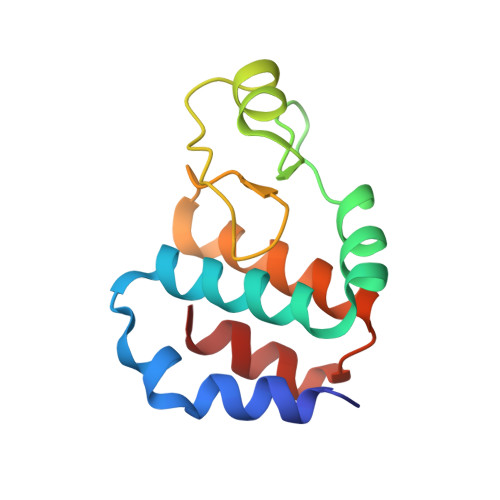
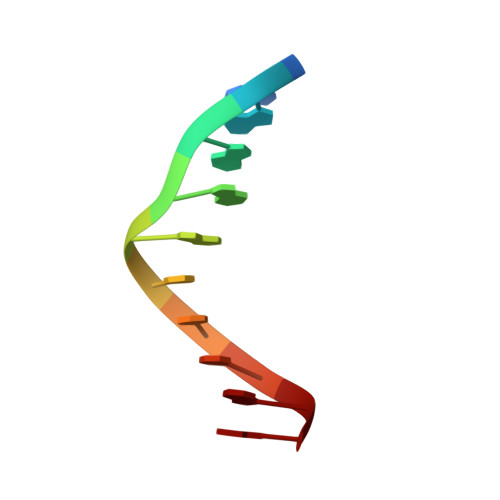
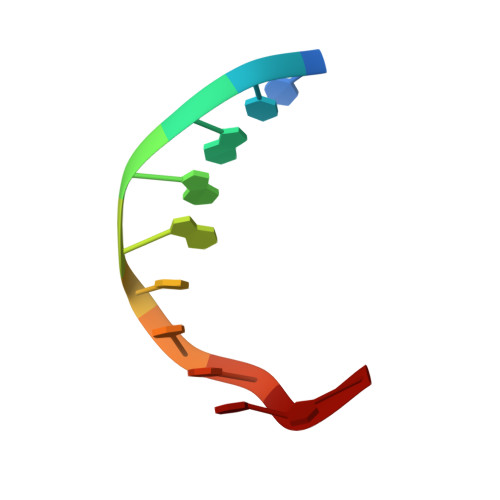
Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein.
Fujikawa, N., Kurumizaka, H., Nureki, O., Tanaka, Y., Yamazoe, M., Hiraga, S., Yokoyama, S.(2004) Nucleic Acids Res 32: 82-92
- PubMed: 14704346
- DOI: https://doi.org/10.1093/nar/gkh173
- Primary Citation of Related Structures:
1IU3, 1J3E - PubMed Abstract:
The Escherichia coli SeqA protein recognizes the 11 hemimethylated G-mA-T-C sites in the oriC region of the chromosome, and prevents replication over-initiation within one cell cycle. The crystal structure of the SeqA C-terminal domain with hemimethylated DNA revealed the N6-methyladenine recognition mechanism; however, the mechanism of discrimination between the hemimethylated and fully methylated states has remained elusive. In the present study, we performed mutational analyses of hemimethylated G-mA-T-C sequences with the minimal DNA-binding domain of SeqA (SeqA71-181), and found that SeqA71-181 specifically binds to hemimethylated DNA containing a sequence with a mismatched mA:G base pair [G-mA(:G)-T-C] as efficiently as the normal hemimethylated G-mA(:T)-T-C sequence. We determined the crystal structures of SeqA71-181 complexed with the mismatched and normal hemimethylated DNAs at 2.5 and 3.0 A resolutions, respectively, and found that the mismatched mA:G base pair and the normal mA:T base pair are recognized by SeqA in a similar manner. Furthermore, in both crystal structures, an electron density is present near the unmethylated adenine, which is only methylated in the fully methylated state. This electron density, which may be due to a water molecule or a metal ion, can exist in the hemimethylated state, but not in the fully methylated state, because of steric clash with the additional methyl group.
- RIKEN Genomic Sciences Center, 1-7-22 Suehiro-cho, Tsurumi, Yokohama 230-0045, Japan.
Organizational Affiliation: