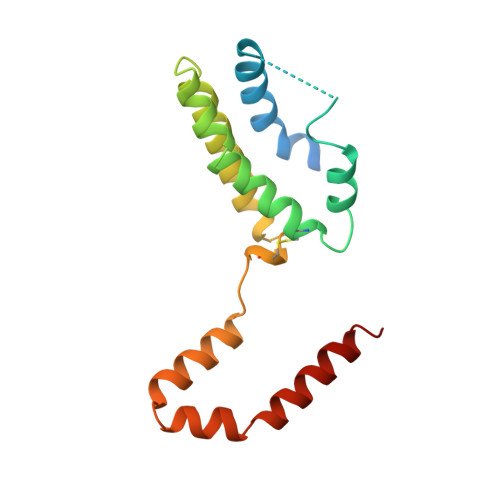
Crystal structure of interleukin 10 reveals an interferon gamma-like fold.
Walter, M.R., Nagabhushan, T.L.(1995) Biochemistry 34: 12118-12125
- PubMed: 7547951
- DOI: https://doi.org/10.1021/bi00038a004
- Primary Citation of Related Structures:
1INR - PubMed Abstract:
The crystal structure of recombinant human interleukin 10 (rhIL-10) has been determined by X-ray crystallography at 2.0 A resolution. Interleukin 10 is a dimer composed of identical polypeptide chains related by a 2-fold axis. The molecule is predominantly alpha-helical. The main-chain fold resembles that of interferon gamma (IFN-gamma) in which the structural integrity of each domain is dependent on the intertwining of helices from each peptide chain. Comparison of rhIL-10 and IFN-gamma reveals differences in helix lengths and orientations of the 2-fold related domains. Interleukin 10 and IFN-gamma contain several conserved residues in their internal cores which suggest a possible "fingerprint" for detection of other members of this fold.
- Department of Pharmacology, University of Alabama at Birmingham 35294, USA.
Organizational Affiliation: