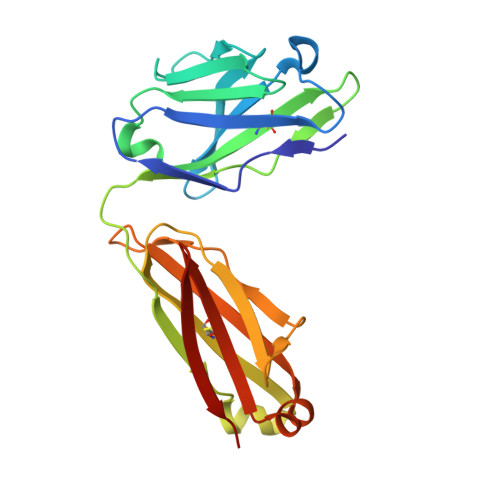
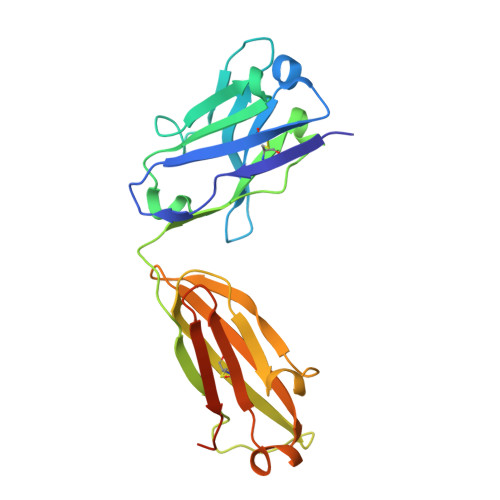
How the anti-(metal chelate) antibody CHA255 is specific for the metal ion of its antigen: X-ray structures for two Fab'/hapten complexes with different metals in the chelate.
Love, R.A., Villafranca, J.E., Aust, R.M., Nakamura, K.K., Jue, R.A., Major Jr., J.G., Radhakrishnan, R., Butler, W.F.(1993) Biochemistry 32: 10950-10959
- PubMed: 8218161
- DOI: https://doi.org/10.1021/bi00092a004
- Primary Citation of Related Structures:
1IND, 1INE - PubMed Abstract:
Antibodies with bound metal-chelate haptens provide new means for exploiting the diverse properties of metallic elements. The murine monoclonal antibody CHA255 (IgG1 lambda) binds the metal-chelate hapten indium (III)-4-[N'-(2-hydroxyethyl)thioureido]-L-benzyl-EDTA (designated In-EOTUBE) with high affinity (K(a) = 1.1 x 10(10) M-1). Antibody binding is highly specific for the indium chelate; the affinity decreases as much as 10(4) with other metals, even those having ionic radii close to indium. To better understand this selectivity, the crystal structure of the antigen-binding fragment (Fab') of CHA255 complexed with its hapten, In(III)-EOTUBE, was determined by molecular replacement and refined at 2.2-A resolution. The structure of CHA255 Fab' complexed with Fe(III)-EOTUBE was also determined and refined at 2.8-A resolution. In both structures, the hapten's EDTA moiety is half-buried near the center of the complementarity-determining regions (CDR's). Five of the six CDR's on the Fab' interact with the hapten through protein side-chain atoms (but not main-chain atoms). A novel feature of the In-EOTUBE/Fab' complex is coordination of the indium by N epsilon of one histidine from the heavy chain's third CDR (distance = 2.4 A). The histidine coordination is not observed in the Fe-EOTUBE/Fab' complex, due mainly to a slightly different hapten conformation that reduces metal accessibility; this may partially explain the 20-fold lower affinity of CHA255 for iron hapten. An unexpected feature of the Fab' overall is an elbow angle of 193 degrees (the angle between the pseudodyad axes of the Fab's constant and variable domains).
- Agouron Pharmaceuticals, Inc., San Diego, California 92121.
Organizational Affiliation: