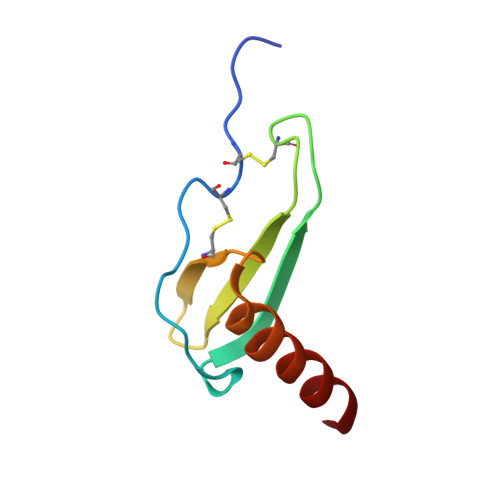
Three-dimensional structure of interleukin 8 in solution.
Clore, G.M., Appella, E., Yamada, M., Matsushima, K., Gronenborn, A.M.(1990) Biochemistry 29: 1689-1696
- PubMed: 2184886
- DOI: https://doi.org/10.1021/bi00459a004
- Primary Citation of Related Structures:
1IL8, 2IL8 - PubMed Abstract:
The solution structure of the interleukin 8 (IL-8) dimer has been solved by nuclear magnetic resonance (NMR) spectroscopy and hybrid distance geometry-dynamical simulated annealing calculations. The structure determination is based on a total of 1880 experimental distance restraints (of which 82 are intersubunit) and 362 torsion angle restraints (comprising phi, psi, and chi 1 torsion angles). A total of 30 simulated annealing structures were calculated, and the atomic rms distribution about the mean coordinate positions (excluding residues 1-5 of each subunit) is 0.41 +/- 0.08 A for the backbone atoms and 0.90 +/- 0.08 A for all atoms. The three-dimensional solution structure of the IL-8 dimer reveals a structural motif in which two symmetry-related antiparallel alpha-helices, approximately 24 A long and separated by about 14 A, lie on top of a six-stranded antiparallel beta-sheet platform derived from two three-stranded Greek keys, one from each monomer unit. The general architecture is similar to that of the alpha 1/alpha 2 domains of the human class I histocompatibility antigen HLA-A2. It is suggested that the two alpha-helices form the binding site for the cellular receptor and that the specificity of IL-8, as well as that of a number of related proteins involved in cell-specific chemotaxis, mediation of cell growth, and the inflammatory response, is achieved by the distinct distribution of charged and polar residues at the surface of the helices.
- Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892.
Organizational Affiliation: