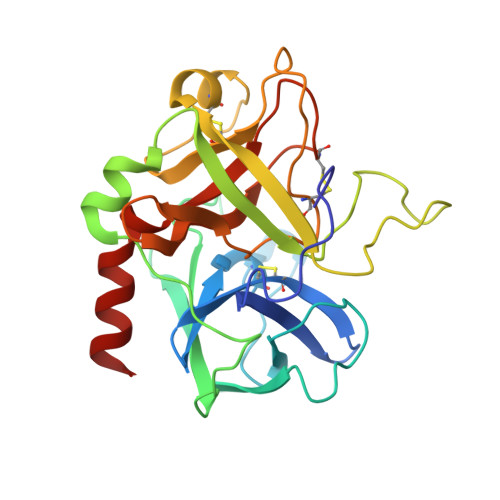
Crystal structure of the complex of human alpha-thrombin and nonhydrolyzable bifunctional inhibitors, hirutonin-2 and hirutonin-6.
Zdanov, A., Wu, S., DiMaio, J., Konishi, Y., Li, Y., Wu, X., Edwards, B.F., Martin, P.D., Cygler, M.(1993) Proteins 17: 252-265
- PubMed: 8272424
- DOI: https://doi.org/10.1002/prot.340170304
- Primary Citation of Related Structures:
1IHS, 1IHT - PubMed Abstract:
The crystal structure of the complexes of hirutonin-2 and hirutonin-6 with human alpha-thrombin have been solved and refined to R-factors of 0.169 (2.0 A resolution) and 0.162 (2.1 A), respectively. Hirutonins belong to a family of bifunctional inhibitors bearing a noncleavable moiety mimicking the scissile bond. Hirutonin-2 is an analog of (D)Phe-Pro-Arg-Gly-hirudin49-65; hirutonin-6 has the same N-terminal tripeptide connected to a shortened fibrinogen exosite-binding part by a short, nonpeptidyl linker. The hirutonin-6 molecule is well defined in the electron density with the exception of the C-terminal Leu-h61. The linker follows near the bottom of the canyon connecting the active site with the exosite, forms a short antiparallel beta-sheet-like arrangement with Leu40-Leu41 and makes van der Waals contacts with Glu39-Leu40-Leu41 of thrombin. In the thrombin-hirutonin-2 complex, the N- and C-terminal parts of the inhibitor are well ordered (except the C-terminal Gln-h65) while the central portion of the linker is partially disordered. The glycine analog in the P1' position of hirutonin-2 assumes a conformation similar to that of the canonical form (Bode and Huber (1992) Eur. J. Biochem. 204:433-451) and supports the identification of the S1' site as restricted by His57, Trp60D, Lys60F, and the Cys42-Cys58 disulfide bridge. The carbonyl oxygen of the P1 arginine residue is located in the oxyanion hole formed by the NH groups of Gly193 and Ser195, while the carbonyl carbon is positioned within a short distance, 2.8 A, from the O gamma of Ser195. This resembles the conformation of the substrate-like inhibitors bound to other serine proteases. The N-terminal (D)Phe-Pro-Arg fragment common to both inhibitors binds to thrombin in a fashion very similar to that of other inhibitors having this motif. The binding of the C-terminus of hirutonins to the fibrinogen-binding exosite is similar to that observed in hirudin and hirulog complexes.
- Biotechnology Research Institute, National Research Council of Canada, Montréal, Québec.
Organizational Affiliation: