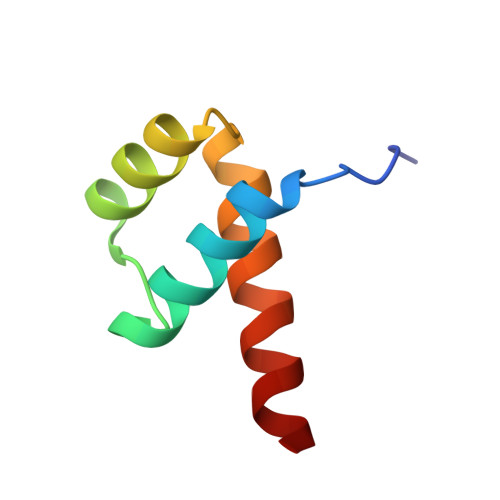
Crystal structure of the Msx-1 homeodomain/DNA complex
Hovde, S., Abate-Shen, C., Geiger, J.H.(2001) Biochemistry 40: 12013-12021
- PubMed: 11580277
- DOI: https://doi.org/10.1021/bi0108148
- Primary Citation of Related Structures:
1IG7 - PubMed Abstract:
The Msx-1 homeodomain protein plays a crucial role in craniofacial, limb, and nervous system development. Homeodomain DNA-binding domains are comprised of 60 amino acids that show a high degree of evolutionary conservation. We have determined the structure of the Msx-1 homeodomain complexed to DNA at 2.2 A resolution. The structure has an unusually well-ordered N-terminal arm with a unique trajectory across the minor groove of the DNA. DNA specificity conferred by bases flanking the core TAAT sequence is explained by well ordered water-mediated interactions at Q50. Most interactions seen at the TAAT sequence are typical of the interactions seen in other homeodomain structures. Comparison of the Msx-1-HD structure to all other high resolution HD-DNA complex structures indicate a remarkably well-conserved sphere of hydration between the DNA and protein in these complexes.
- Michigan State University Chemistry Department, East Lansing Michigan 48824, USA.
Organizational Affiliation: