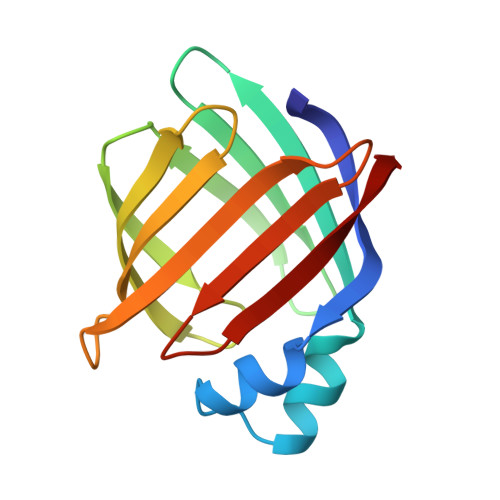
Refinement of the structure of recombinant rat intestinal fatty acid-binding apoprotein at 1.2-A resolution.
Scapin, G., Gordon, J.I., Sacchettini, J.C.(1992) J Biological Chem 267: 4253-4269
- PubMed: 1740465
- DOI: https://doi.org/10.2210/pdb1ifc/pdb
- Primary Citation of Related Structures:
1IFC - PubMed Abstract:
The three-dimensional structure of the 131-residue rat intestinal fatty acid-binding protein, without bound ligand (apoI-FABP), has been refined with x-ray diffraction data to a nominal resolution of 1.19 A. The final model has a conventional crystallographic R-factor of 16.9% for 34,290 unique reflections [a root mean square (r.m.s.) deviation for bond length of 0.012 A and a r.m.s. deviation of 2.368 degrees for bond angles]. Ninety-two residues are present as components of the protein's 10 anti-parallel beta-strands while 14 residues are part of its two short alpha-helices. The beta-strands and alpha-helices are organized into two nearly orthogonal beta-sheets. Particular attention has been placed in defining solvent structure and the structures of discretely disordered groups in this protein. Two hundred thirty-seven solvent molecules have been identified; 24 are located within apoI-FABP. The refined model includes alternate conformers for 228 protein atoms (109 main-chain, 119 side-chain) and 63 solvent molecules. We have found several aromatic side-chains with multiple conformations located near, or in, the protein's ligand binding site. This observation, along with the fact that these side-chains have a temperature factor that is relatively higher than that of other aromatic residues, suggests that they may be involved in the process of noncovalent binding of fatty acid. The absence of a true hydrophobic core in I-FABP suggests that its structural integrity may be maintained primarily by a hydrogen bonding network involving protein and solvent atoms.
- Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461.
Organizational Affiliation: