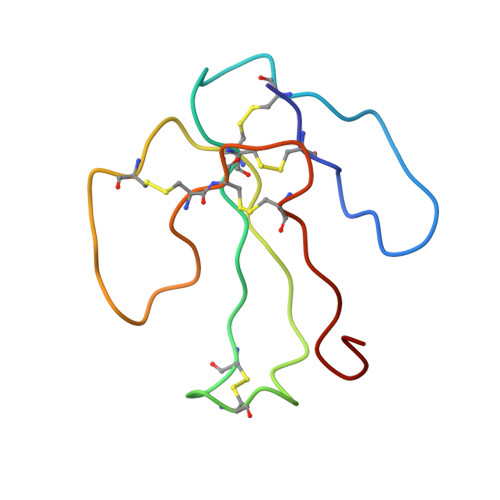
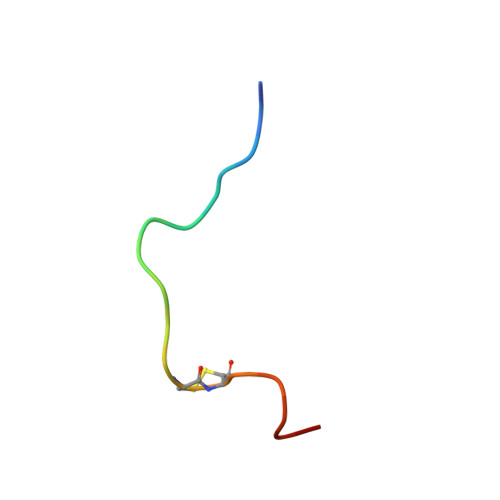
The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
Zeng, H., Moise, L., Grant, M.A., Hawrot, E.(2001) J Biological Chem 276: 22930-22940
- PubMed: 11312275
- DOI: https://doi.org/10.1074/jbc.M102300200
- Primary Citation of Related Structures:
1IDG, 1IDH, 1IDI, 1IDL - PubMed Abstract:
The region encompassing residues 181-98 on the alpha1 subunit of the muscle-type nicotinic acetylcholine receptor forms a major determinant for the binding of alpha-neurotoxins. We have prepared an (15)N-enriched 18-amino acid peptide corresponding to the sequence in this region to facilitate structural elucidation by multidimensional NMR. Our aim was to determine the structural basis for the high affinity, stoichiometric complex formed between this cognate peptide and alpha-bungarotoxin, a long alpha-neurotoxin. Resonances in the complex were assigned through heteronuclear and homonuclear NMR experiments, and the resulting interproton distance constraints were used to generate ensemble structures of the complex. Thr(8), Pro(10), Lys(38), Val(39), Val(40), and Pro(69) in alpha-bungarotoxin and Tyr(189), Tyr(190), Thr(191), Cys(192), Asp(195), and Thr(196) in the peptide participate in major intermolecular contacts. A comparison of the free and bound alpha-bungarotoxin structures reveals significant conformational rearrangements in flexible regions of alpha-bungarotoxin, mainly loops I, II, and the C-terminal tail. Furthermore, several of the calculated structures suggest that cation-pi interactions may be involved in binding. The root mean square deviation of the polypeptide backbone in the complex is 2.07 A. This structure provides, to date, the highest resolution description of the contacts between a prototypic alpha-neurotoxin and its cognate recognition sequence.
- Department of Molecular Pharmacology, Physiology, and Biotechnology, Brown Medical School, Providence, Rhode Island 02912, USA.
Organizational Affiliation: