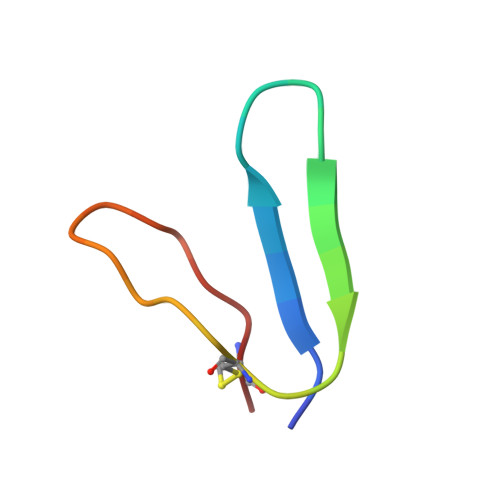
Design of a discretely folded mini-protein motif with predominantly beta-structure.
Ottesen, J.J., Imperiali, B.(2001) Nat Struct Biol 8: 535-539
- PubMed: 11373623
- DOI: https://doi.org/10.1038/88604
- Primary Citation of Related Structures:
1IC9, 1ICL, 1ICO - PubMed Abstract:
Here we report the creation of a predominantly beta-structured mini-protein motif. The design target is based on the naturally occurring toxin hand (TH) motifs that are composed of four disulfide bonds and three loops that form a 'hand'. Analysis and subsequent modification of several generations of mini-proteins produced the final 29-residue mini-protein. The structured motif of this new mini-protein provides insight into the compensatory changes that result in the formation of a tightly packed hydrophobic core in a small, globular beta-structure motif. Additionally, this mini-motif represents a new, distinct surface topology for protein design and a valuable, yet compact, model system for the study of beta-sheet structure in water.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.
Organizational Affiliation: