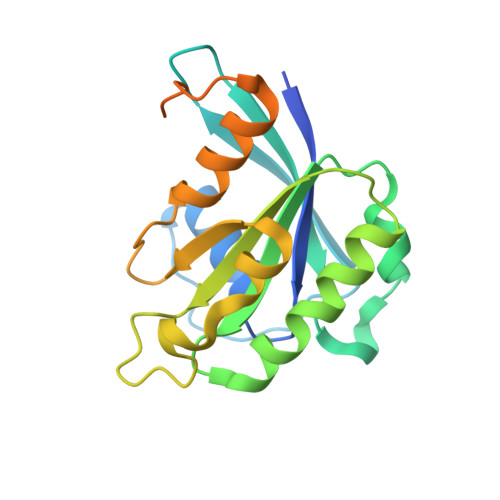
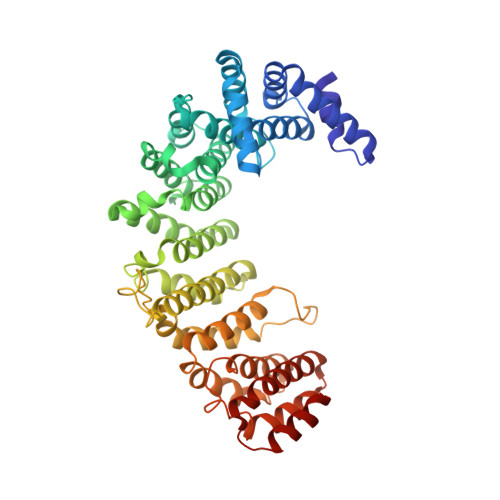
Structural view of the Ran-Importin beta interaction at 2.3 A resolution
Vetter, I.R., Arndt, A., Kutay, U., Gorlich, D., Wittinghofer, A.(1999) Cell 97: 635-646
- PubMed: 10367892
- DOI: https://doi.org/10.1016/s0092-8674(00)80774-6
- Primary Citation of Related Structures:
1IBR - PubMed Abstract:
Transport receptors of the Importin beta family shuttle between the nucleus and cytoplasm and mediate transport of macromolecules through nuclear pore complexes. They interact specifically with the GTP-binding protein Ran, which in turn regulates their interaction with cargo. Here, we report the three-dimensional structure of a complex between Ran bound to the nonhydrolyzable GTP analog GppNHp and a 462-residue fragment from Importin beta. The structure of Importin beta shows 10 tandem repeats resembling HEAT and Armadillo motifs. They form an irregular crescent, the concave site of which forms the interface with Ran-triphosphate. The importin-binding site of Ran does not overlap with that of the Ran-binding domain of RanBP2.
- Max-Planck-Institut für molekulare Physiologie, Dortmund, Germany.
Organizational Affiliation: