SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE
Yu, L., Gunasekera, A.H., Mack, J., Olejniczak, E.T., Chovan, L.E., Ruan, X., Towne, D.L., Lerner, C.G., Fesik, S.W.(2001) J Mol Biology 311: 593-604
- PubMed: 11493012
- DOI: https://doi.org/10.1006/jmbi.2001.4894
- Primary Citation of Related Structures:
1IB8 - PubMed Abstract:
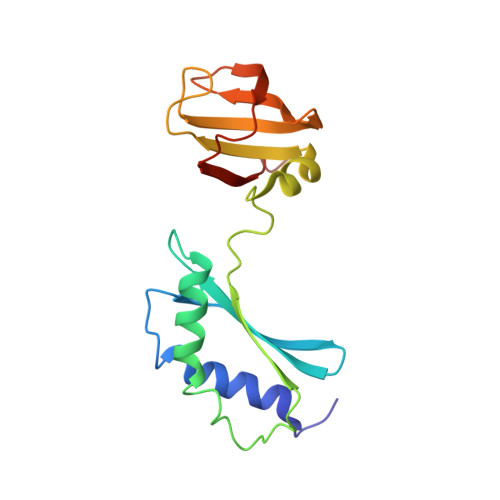
Streptococcus pneumoniae is a major human pathogen that causes high mortality and morbidity rates and has developed resistance to many antibiotics. The genome of S. pneumoniae has recently been completely sequenced revealing many genes encoding hypothetical proteins of unknown function. We have found that the gene encoding one such conserved protein, SP14.3, is essential for growth of S. pneumonia. Since it is essential, SP14.3 represents a potential target for drug discovery. Here, we describe the three-dimensional solution structure of SP14.3 as determined by NMR spectroscopy. The structure consists of two domains each with an alpha/beta-fold. The N-terminal domain contains two alpha-helices and a three-stranded beta-sheet, while the C-terminal domain is composed of one alpha-helix and a five-stranded beta-sheet. The N-terminal domain of the protein contains a highly negatively charged surface and resembles the fold of the N-terminal domain of Thermus thermophilus ribosomal protein S3. The C-terminal domain has a protein fold similar to human small nuclear ribonucleoprotein Sm D3 and Haloarcula marismortui ribosomal protein L21E. The two domains of the protein tumble in solution overall as a whole with an overall molecular rotational correlation time (tau(m)) of 12.9 ns at 25 degrees C. The relative orientation of the two domains is not defined by the nuclear Overhauser effect data. Indeed, residual dipolar couplings and the structure calculations indicate that the relative orientation of the two domains is not rigidly oriented with respect to one another in solution.
- Pharmaceutical Discovery Division, Abbott Park, IL, 60064-6098, USA. liping.yu@abbott.com
Organizational Affiliation: