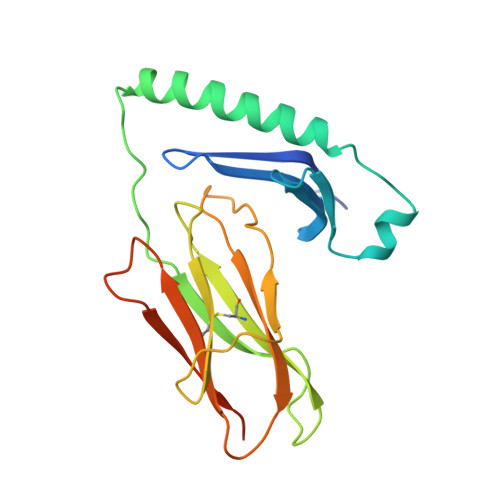
Crystal structure of I-Ak in complex with a dominant epitope of lysozyme.
Fremont, D.H., Monnaie, D., Nelson, C.A., Hendrickson, W.A., Unanue, E.R.(1998) Immunity 8: 305-317
- PubMed: 9529148
- DOI: https://doi.org/10.1016/s1074-7613(00)80536-1
- Primary Citation of Related Structures:
1IAK - PubMed Abstract:
We have determined the structure of murine MHC class II I-Ak in complex with a naturally processed peptide from hen egg lysozyme (HEL residues 50-62) at 1.9 A resolution. These results provide a structural basis for the I-Ak peptide-binding motif. Binding is established by the deep burial of five anchor side chains into specific pockets of the I-Ak binding groove, with a zen-like fit of an aspartic acid in the P1 pocket. We also show that in the I-Ak alpha chain, a bulge occurs in the first strand of the peptide-binding platform, an insertion probably common to all I-A and HLA-DQ alleles. The I-Ak beta chain has a deletion in the helical region adjacent to the P7 pocket and an insertion in the helical region neighboring the P1 pocket. As a result of these structural features, the extended HEL peptide dips low into the center of the I-Ak groove and reaches toward solvent at its C-terminal end.
- Howard Hughes Medical Institute, Department of Biochemistry and Molecular Biophysics, Columbia University, New York, New York 10032, USA. fremont@immunology.wustl.edu
Organizational Affiliation: