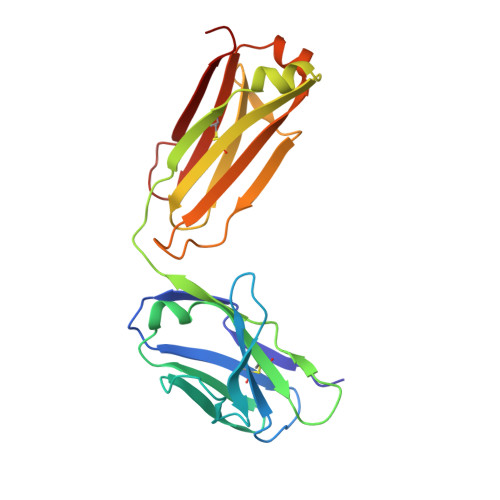
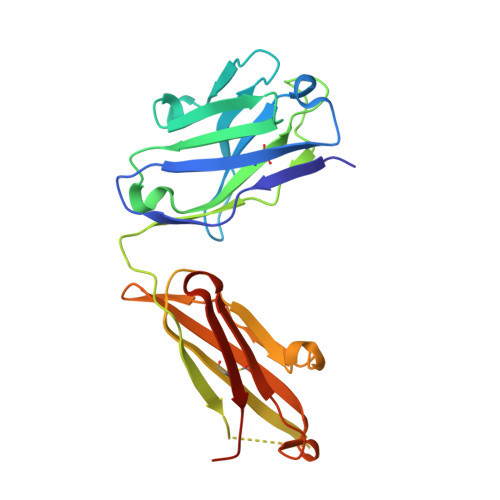
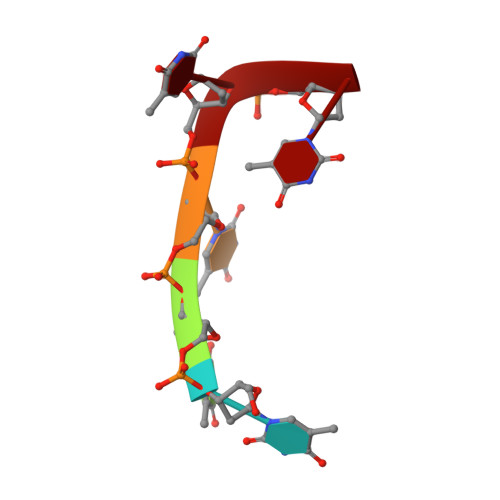
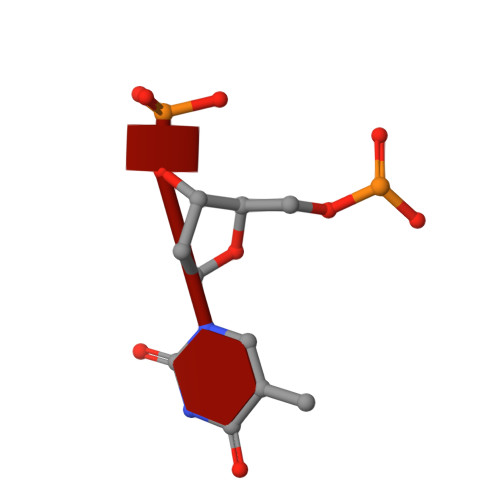
Crystal Structure of an Antigen-Binding Fragment Bound to Single-Stranded DNA
Tanner, J.J., Komissarov, A.A., Deutscher, S.L.(2001) J Mol Biology 314: 807-822
- PubMed: 11733999
- DOI: https://doi.org/10.1006/jmbi.2001.5178
- Primary Citation of Related Structures:
1I8M - PubMed Abstract:
Antibodies to DNA are characteristic of the autoimmune disease systemic lupus erythematosus (SLE) and they also serve as models for the study of protein-DNA recognition. Anti-DNA antibodies often play an important role in disease pathogenesis by mediating kidney damage via antibody-DNA immune complex formation. The structural underpinnings of anti-DNA antibody pathogenicity and antibody-DNA recognition, however, are not well understood, due in part to the lack of direct, experimental three-dimensional structural information on antibody-DNA complexes. To address these issues for anti-single-stranded DNA antibodies, we have determined the 2.1 A crystal structure of a recombinant Fab (DNA-1) in complex with dT5. DNA-1 was previously isolated from a bacteriophage Fab display library from the immunoglobulin repertoire of an SLE-prone mouse. The structure shows that DNA-1 binds oligo(dT) primarily by sandwiching thymine bases between Tyr side-chains, which allows the bases to make sequence-specific hydrogen bonds. The critical stacking Tyr residues are L32, L49, H100, and H100A, while His L91 and Asn L50 contribute hydrogen bonds. Comparison of the DNA-1 structure to other anti-nucleic acid Fab structures reveals a common ssDNA recognition module consisting of Tyr L32, a hydrogen bonding residue at position L91, and an aromatic side-chain from the tip of complementarity determining region H3. The structure also provides a framework for interpreting previously determined thermodynamics data, and this analysis suggests that hydrophobic desolvation might underlie the observed negative enthalpy of binding. Finally, Arg side-chains from complementarity determining region H3 appear to play a novel role in DNA-1. Rather than forming ion pairs with dT5, Arg contributes to oligo(dT) recognition by helping to maintain the structural integrity of the combining site. This result is significant because antibody pathogenicity is thought to be correlated to the Arg content of anti-DNA antibody hypervariable loops.
- Department of Chemistry, University of Missouri-Columbia, Columbia, MO 65211, USA. tannerjj@missouri.edu
Organizational Affiliation: