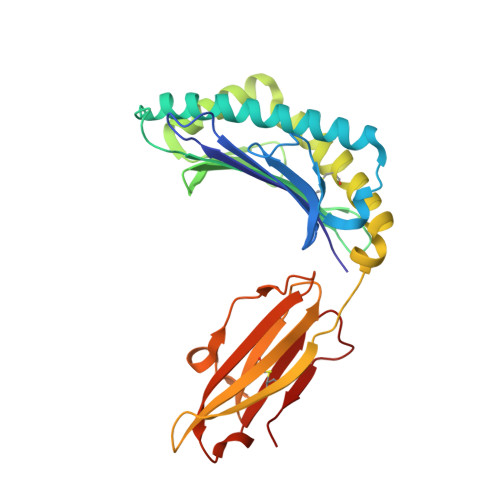
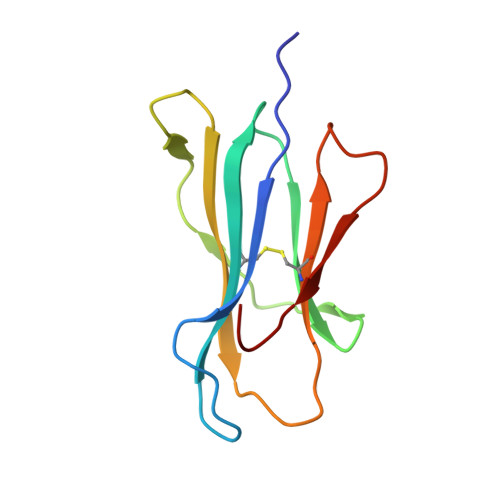
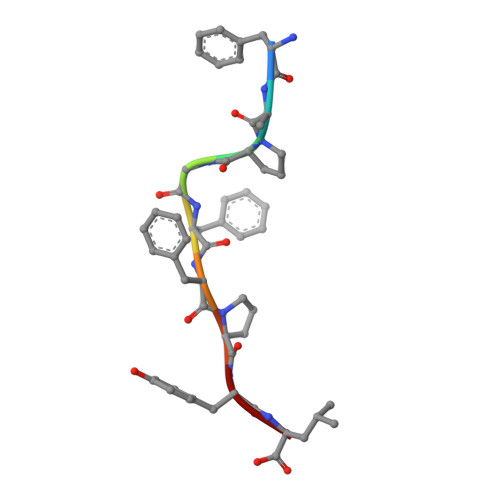
T cell activity correlates with oligomeric peptide/MHC binding on T cell surface
Busslep, J., Zhao, R., Donnini, D., Loftus, D., Saad, M., Appella, E., Collins, E.J.(2001) J Biological Chem 276: 47320-47328
- PubMed: 11584024
- DOI: https://doi.org/10.1074/jbc.M109231200
- Primary Citation of Related Structures:
1I7R, 1I7T, 1I7U - PubMed Abstract:
Recognition of virally infected cells by CD8+ T cells requires differentiation between self and nonself peptide-class I major histocompatibility complexes (pMHC). Recognition of foreign pMHC by host T cells is a major factor in the rejection of transplanted organs from the same species (allotransplant) or different species (xenotransplant). AHIII12.2 is a murine T cell clone that recognizes the xenogeneic (human) class I MHC HLA-A2.1 molecule (A2) and the syngeneic murine class I MHC H-2 D(b) molecule (D(b)). Recognition of both A2 and D(b) are peptide-dependent, and the sequences of the peptides recognized have been determined. Alterations in the antigenic peptides bound to A2 cause large changes in AHIII12.2 T cell responsiveness. Crystal structures of three representative peptides (agonist, null, and antagonist) bound to A2 partially explain the changes in AHIII12.2 responsiveness. Using class I pMHC octamers, a strong correlation is seen between T cell activity and the affinity of pMHC complexes for the T cell receptor. However, contrary to previous studies, we see similar half-lives for the pMHC multimers bound to the AHIII12.2 cell surface.
- Department of Microbiology and Immunology, University of North Carolina, Chapel Hill, NC 27599, USA.
Organizational Affiliation: