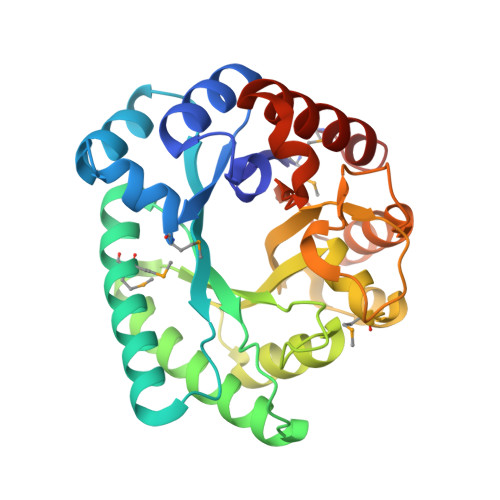
Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Zhang, R.G., Dementieva, I., Duke, N., Collart, F., Quaite-Randall, E., Alkire, R., Dieckman, L., Maltsev, N., Korolev, O., Joachimiak, A.(2002) Proteins 48: 423-426
- PubMed: 12112707
- DOI: https://doi.org/10.1002/prot.10159
- Primary Citation of Related Structures:
1I60, 1I6N - Biosciences Division, Structural Biology Center, Argonne National Laboratory, Argonne, Illinois 60439, USA.
Organizational Affiliation: