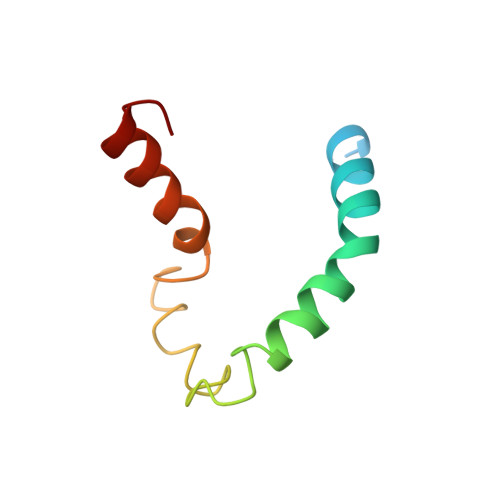
NMR structure of human apolipoprotein C-II in the presence of sodium dodecyl sulfate.
MacRaild, C.A., Hatters, D.M., Howlett, G.J., Gooley, P.R.(2001) Biochemistry 40: 5414-5421
- PubMed: 11331005
- DOI: https://doi.org/10.1021/bi002821m
- Primary Citation of Related Structures:
1I5J - PubMed Abstract:
The structure and protein-detergent interactions of apolipoprotein C-II (apoC-II) in the presence of SDS micelles have been investigated using circular dichroism and heteronuclear NMR techniques applied to (15)N-labeled protein. Micellar SDS, a commonly used mimetic of the lipoprotein surface, inhibits the aggregation of apoC-II and induces a stable structure containing approximately 60% alpha-helix as determined by circular dichroism. NMR reveals the first 12 residues of apoC-II to be structurally heterogeneous and largely disordered, with the rest of the protein forming a predominantly helical structure. Three regions of helical conformation, residues 16-36, 50-56, and 63-77, are well-defined by NMR-derived constraints, with the intervening regions showing more loosely defined helical conformation. The structure of apoC-II is compared to that determined for other apolipoproteins in a similar environment. Our results shed light on the lipid interactions of apoC-II and its mechanism of lipoprotein lipase activation.
- Russell Grimwade School of Biochemistry and Molecular Biology, University of Melbourne, Gate 12, Royal Parade, Parkville, Victoria 3052, Australia.
Organizational Affiliation: