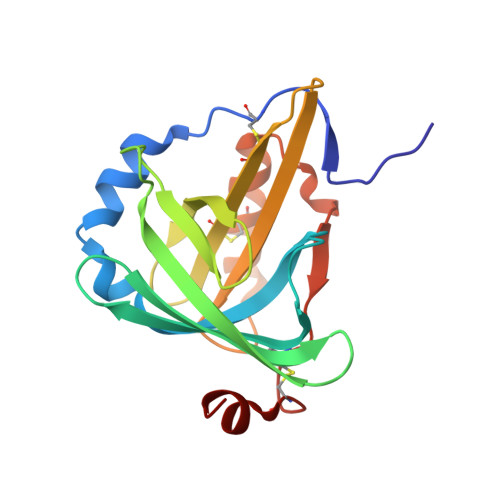
The C1 subunit of alpha-crustacyanin: the de novo phasing of the crystal structure of a 40 kDa homodimeric protein using the anomalous scattering from S atoms combined with direct methods.
Gordon, E.J., Leonard, G.A., McSweeney, S., Zagalsky, P.F.(2001) Acta Crystallogr D Biol Crystallogr 57: 1230-1237
- PubMed: 11526314
- DOI: https://doi.org/10.1107/s0907444901009362
- Primary Citation of Related Structures:
1I4U - PubMed Abstract:
The previously unknown crystal structure of the C(1) subunit of the carotenoid-binding protein alpha-crustacyanin has been determined using the anomalous scattering available at 1.77 A wavelength to determine the partial structure of the S atoms intrinsic to the native protein. The resulting 'heavy-atom' phases, in conjunction with near-atomic resolution (d(min) = 1.15 A) data, were then used to initiate successful structure determination using a direct-methods approach. This is, to the authors' knowledge, the first time such a small anomalous signal ( approximately 1%) has been used to aid the determination of a macromolecular structure. As well as the structure itself, the methods used during data collection and those used in the elucidation of the sulfur 'heavy-atom' partial structure are described here. As predicted, the C(1) subunit adopts a tertiary structure typical of the lipocalin superfamily: an eight-stranded antiparallel beta-barrel with a repeated +1 topology. The beta-barrel has a calyx shape with the two molecules in the asymmetric unit interacting in such a way that the open ends of each calyx face each other, although they do not form a single elongated pocket. A comparison of this structure with those of other members of the lipocalin superfamily has allowed speculation as to the nature of carotenoid binding by the protein.
- Macromolecular Crystallography, European Synchrotron Radiation Facility, BP 220, F-38043 Grenoble CEDEX, France. gordon@esrf.fr
Organizational Affiliation: