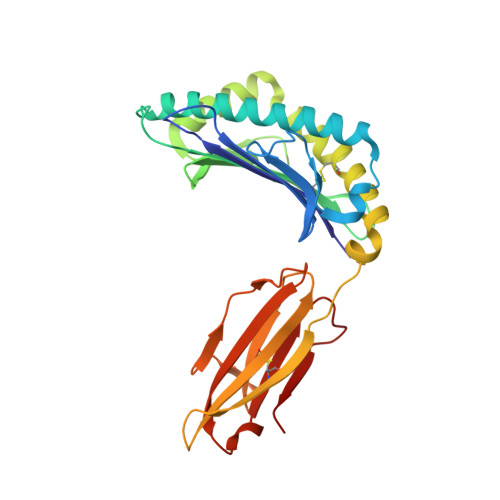
High-resolution structure of HLA-A*0201 in complex with a tumour-specific antigenic peptide encoded by the MAGE-A4 gene.
Hillig, R.C., Coulie, P.G., Stroobant, V., Saenger, W., Ziegler, A., Hulsmeyer, M.(2001) J Mol Biology 310: 1167-1176
- PubMed: 11502003
- DOI: https://doi.org/10.1006/jmbi.2001.4816
- Primary Citation of Related Structures:
1I4F - PubMed Abstract:
The heterotrimeric complex of the human major histocompatibity complex (MHC) molecule HLA-A*0201, beta2-microglobulin and the decameric peptide GVYDGREHTV derived from the melanoma antigen (MAGE-A4 protein has been determined by X-ray crystallography at 1.4 A resolution. MAGE-A4 belongs to a family of genes that are specifically expressed in a variety of tumours. MAGE-A4-derived peptides are presented by MHC molecules at the cell surface to cytotoxic T-lymphocytes. As the HLA-A*0201:MAGE-A4 complex occurs only on tumour cells, it is considered to be an appropriate target for immunotherapy. The structure presented here reveals potential epitopes specific to the complex and indicates which peptide residues could be recognised by T-cell receptors. In addition, as the structure could be refined anisotropically, it was possible to describe the movements of the bound peptide in more detail.
- Institut für Immungenetik, Universitätsklinikum Charité, Humboldt-Universität zu Berlin, Germany.
Organizational Affiliation: