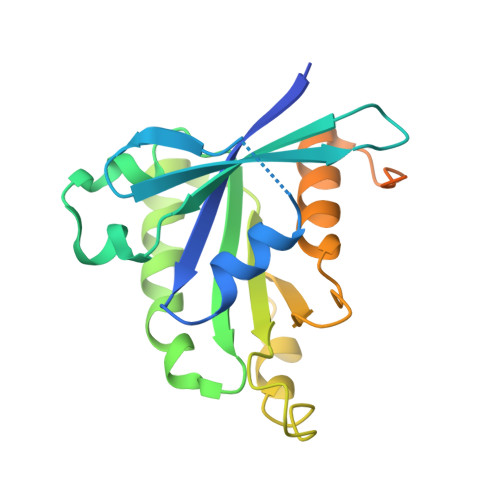
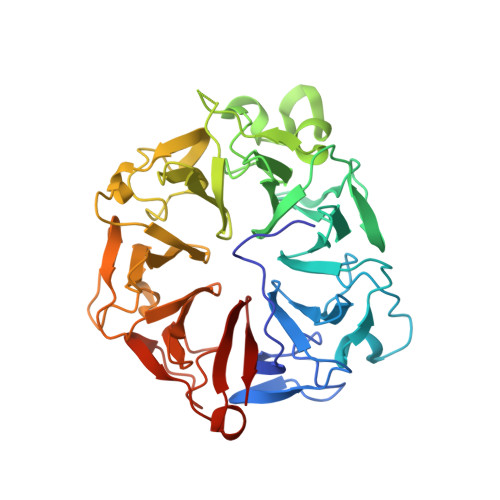
Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1).
Renault, L., Kuhlmann, J., Henkel, A., Wittinghofer, A.(2001) Cell 105: 245-255
- PubMed: 11336674
- DOI: https://doi.org/10.1016/s0092-8674(01)00315-4
- Primary Citation of Related Structures:
1I2M - PubMed Abstract:
RCC1 (regulator of chromosome condensation), a beta propeller chromatin-bound protein, is the guanine nucleotide exchange factor (GEF) for the nuclear GTP binding protein Ran. We report here the 1.8 A crystal structure of a Ran*RCC1 complex in the absence of nucleotide, an intermediate in the multistep GEF reaction. In contrast to previous structures, the phosphate binding region of the nucleotide binding site is perturbed only marginally, possibly due to the presence of a polyvalent anion in the P loop. Biochemical experiments show that a sulfate ion stabilizes the Ran*RCC1 complex and inhibits dissociation by guanine nucleotides. Based on the available structural and biochemical evidence, we present a unified scenario for the GEF mechanism where interaction of the P loop lysine with an acidic residue is a crucial element for the overall reaction.
- Max-Planck-Institut für Molekulare Physiologie, Postfach 50 02 47, 44202, Dortmund, Germany.
Organizational Affiliation: