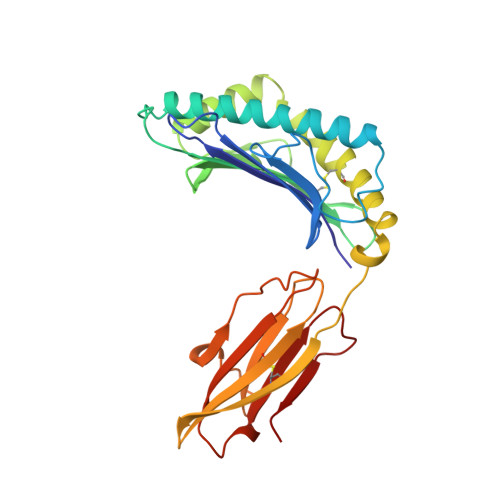
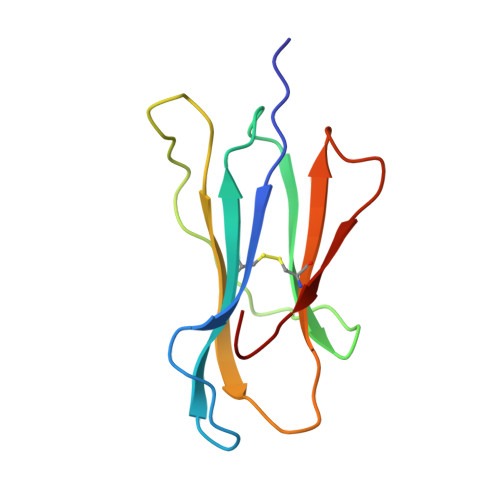
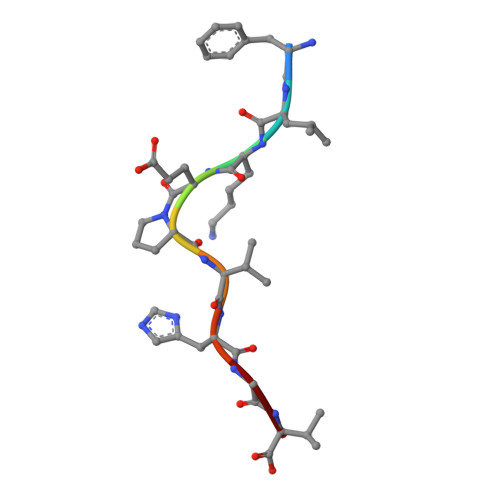
The structural basis for the increased immunogenicity of two HIV-reverse transcriptase peptide variant/class I major histocompatibility complexes.
Kirksey, T.J., Pogue-Caley, R.R., Frelinger, J.A., Collins, E.J.(1999) J Biological Chem 274: 37259-37264
- PubMed: 10601290
- DOI: https://doi.org/10.1074/jbc.274.52.37259
- Primary Citation of Related Structures:
1I1F, 1I1Y - PubMed Abstract:
Designing altered peptide ligands to generate specific immunological reactivity when bound to class I major histocompatibility complexes is important for both therapeutic and prophylactic reasons. We have previously shown that two altered peptides, derived from human immunodeficiency virus (HIV)-reverse transcriptase (RT) residues 309-317, are more immunogenic in vitro than the wild-type peptide. One peptide variant, I1Y, was able to stimulate RT-specific cytotoxic T cells from the blood of three HIV-infected individuals better than the wild-type RT peptide. Both I1Y and I1F peptide variants increase the cell surface half-life of the peptide-class I complex approximately 3-fold over that of the RT peptide but have different immunological activities. These peptides are candidates for the design of vaccines for HIV due to their increased immunogenicity. To understand the basis for the increased cell surface stability compared with wild-type peptide and to understand the differences in T cell recognition between I1Y and I1F, we determined the x-ray crystal structures of the two class I MHC-peptide complexes. These structures indicate that the increased cell surface half-life is due to pi-pi stacking interactions between Trp-167 of HLA-A2.1 and the aromatic P1 residues of I1F and I1Y. Comparison of the structures and modeling potential T cell receptor (TCR) interactions suggests that T cell interactions and immunogenicity are different between I1Y and I1F for two reasons. First, subtle changes in the steric and polar properties of the I1Y peptide affect TCR engagement. Second, water-mediated hydrogen bond interactions between the P1-Tyr and the P4-Glu peptide residues increase peptide side chain rigidity of residues critical for TCR engagement.
- Department of Microbiology, the University of North Carolina, Chapel Hill, North Carolina 27599, USA.
Organizational Affiliation: