The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity.
Musil, D., Zucic, D., Turk, D., Engh, R.A., Mayr, I., Huber, R., Popovic, T., Turk, V., Towatari, T., Katunuma, N., Bode, W.(1991) EMBO J 10: 2321-2330
- PubMed: 1868826
- DOI: https://doi.org/10.1002/j.1460-2075.1991.tb07771.x
- Primary Citation of Related Structures:
1HUC - PubMed Abstract:
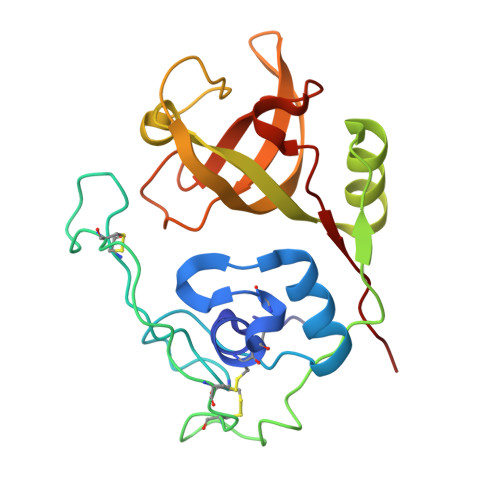
From the lysosomal cysteine proteinase cathepsin B, isolated from human liver in its two-chain form, monoclinic crystals were obtained which contain two molecules per asymmetric unit. The molecular structure was solved by a combination of Patterson search and heavy atom replacement methods (simultaneously with rat cathepsin B) and refined to a crystallographic R value of 0.164 using X-ray data to 2.15 A resolution. The overall folding pattern of cathepsin B and the arrangement of the active site residues are similar to the related cysteine proteinases papain, actinidin and calotropin DI. 166 alpha-carbon atoms out of 248 defined cathepsin B residues are topologically equivalent (with an r.m.s. deviation of 1.04 A) with alpha-carbon atoms of papain. However, several large insertion loops are accommodated on the molecular surface and modify its properties. The disulphide connectivities recently determined for bovine cathepsin B by chemical means were shown to be correct. Some of the primed subsites are occluded by a novel insertion loop, which seems to favour binding of peptide substrates with two residues carboxy-terminal to the scissile peptide bond; two histidine residues (His110 and His111) in this "occluding loop' provide positively charged anchors for the C-terminal carboxylate group of such polypeptide substrates. These structural features explain the well-known dipeptidyl carboxypeptidase activity of cathepsin B. The other subsites adjacent to the reactive site Cys29 are relatively similar to papain; Glu245 in the S2 subsite favours basic P2-side chains. The above mentioned histidine residues, but also the buried Glu171 might represent the group with a pKa of approximately 5.5 near the active site, which governs endo- and exopeptidase activity. The "occluding loop' does not allow cystatin-like protein inhibitors to bind to cathepsin B as they do to papain, consistent with the reduced affinity of these protein inhibitors for cathepsin B compared with the related plant enzymes.
- Max-Planck-Institut für Biochemie, Martinsried, FRG.
Organizational Affiliation: