Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle.
Batey, R.T., Sagar, M.B., Doudna, J.A.(2001) J Mol Biology 307: 229-246
- PubMed: 11243816
- DOI: https://doi.org/10.1006/jmbi.2000.4454
- Primary Citation of Related Structures:
1HQ1 - PubMed Abstract:
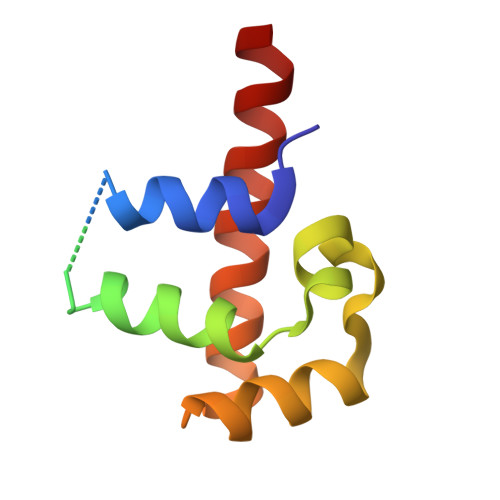
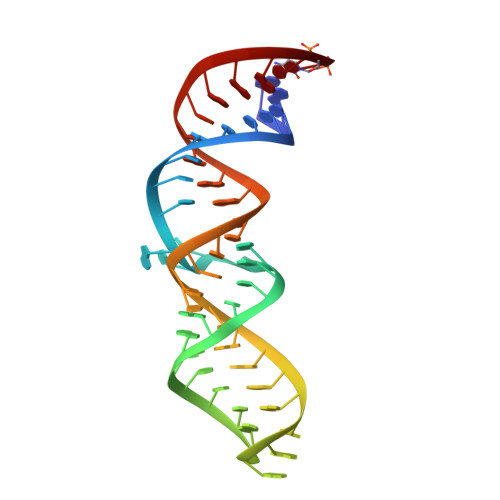
The signal recognition particle (SRP) is a ribonucleoprotein complex responsible for targeting proteins to the endoplasmic reticulum in eukarya or to the inner membrane in prokarya. The crystal structure of the universally conserved RNA-protein core of the Escherichia coli SRP, refined here to 1.5 A resolution, revealed minor groove recognition of the 4.5 S RNA component by the M domain of the Ffh protein. Within the RNA, nucleotides comprising two phylogenetically conserved internal loops create a unique surface for protein recognition. To determine the energetic importance of conserved nucleotides for SRP assembly, we measured the affinity of the M domain for a series of RNA mutants. This analysis reveals how conserved nucleotides within the two internal loop motifs establish the architecture of the macromolecular interface and position essential functional groups for direct recognition by the protein.
- Department of Molecular Biophysics and Biochemistry and Howard Hughes Medical Institute, Yale University, P.O. Box 208114, New Haven, CT 06520-8814, USA.
Organizational Affiliation: