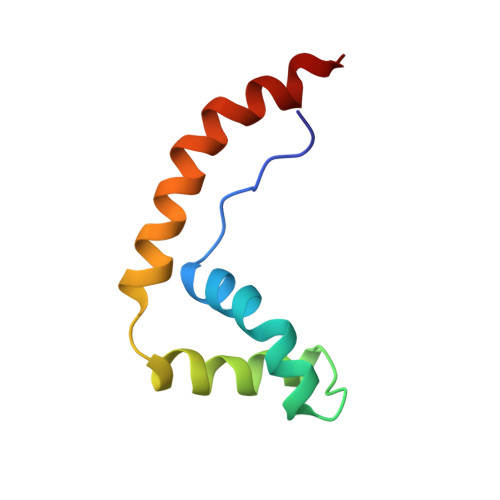
The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Jones, D.N., Searles, M.A., Shaw, G.L., Churchill, M.E., Ner, S.S., Keeler, J., Travers, A.A., Neuhaus, D.(1994) Structure 2: 609-627
- PubMed: 7922039
- DOI: https://doi.org/10.1016/s0969-2126(00)00063-0
- Primary Citation of Related Structures:
1HMA - PubMed Abstract:
The HMG-box is a conserved DNA-binding motif that has been identified in many high mobility group (HMG) proteins. HMG-D is a non-histone chromosomal protein from Drosophila melanogaster that is closely related to the mammalian HMG-box proteins HMG-1 and HMG-2. Previous structures determined for an HMG-box domain from rat and hamster exhibit the same global topology, but differ significantly in detail. It has been suggested that these differences may arise from hinge motions which allow the protein to adapt to the shape of its target DNA. We present the solution structure of HMG-D determined by NMR spectroscopy to an overall precision of 0.85 A root mean squared deviation (rmsd) for the backbone atoms. The protein consists of an extended amino-terminal region and three alpha-helices that fold into a characteristic 'L' shape. The central core region of the molecule is highly stable and maintains an angle of approximately 80 degrees between the axes of helices 2 and 3. The backbone dynamics determined from 15N NMR relaxation measurements show a high correlation with the mean residue rmsd determined from the calculated structures. The structure determined for the HMG-box motif from HMG-D is essentially identical to the structure determined for the B-domain of mammalian HMG-1. Since these proteins have significantly different sequences our results indicate that the global fold and the mode of interaction with DNA are also likely to be conserved in all eukaryotes.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: