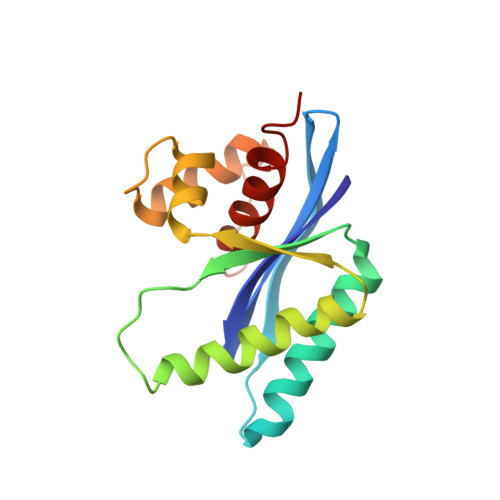
Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli.
Ariyoshi, M., Vassylyev, D.G., Iwasaki, H., Nakamura, H., Shinagawa, H., Morikawa, K.(1994) Cell 78: 1063-1072
- PubMed: 7923356
- DOI: https://doi.org/10.1016/0092-8674(94)90280-1
- Primary Citation of Related Structures:
1HJR - PubMed Abstract:
The crystal structure of the RuvC protein, a Holliday junction resolvase from E. coli, has been determined at 2.5 A resolution. The enzyme forms a dimer of 19 kDa subunits related by a dyad axis. Together with results from extensive mutational analyses, the refined structure reveals that the catalytic center, comprising four acidic residues, lies at the bottom of a cleft that nicely fits a DNA duplex. The structural features of the dimer, with a 30 A spacing between the two catalytic centers, provide a substantially defined image of the Holliday junction architecture. The folding topology in the vicinity of the catalytic site exhibits a striking similarity to that of RNAase H1 from E. coli.
- Protein Engineering Research Institute, Osaka, Japan.
Organizational Affiliation: