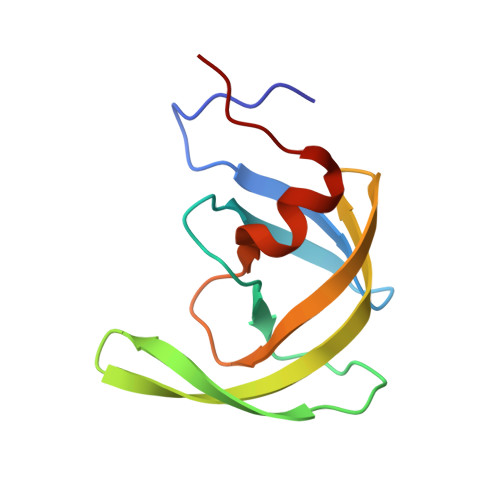
The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU.
Spinelli, S., Liu, Q.Z., Alzari, P.M., Hirel, P.H., Poljak, R.J.(1991) Biochimie 73: 1391-1396
- PubMed: 1799632
- DOI: https://doi.org/10.1016/0300-9084(91)90169-2
- Primary Citation of Related Structures:
1HHP - PubMed Abstract:
The crystal structure of the aspartyl protease encoded by the gene pol of the human immunodeficiency virus (HIV-1, isolate BRU) has been determined to 2.7 A resolution. The enzyme, expressed as an insoluble denatured polypeptide in inclusion bodies of Escherichia coli has been renatured and crystallized. It differs by several amino acid replacements from the homologous enzymes of other HIV-1 isolates. A superposition of the C alpha-backbone of the BRU protease with that of the SF2 protease gives a roots mean square positional difference of 0.45 A. Thus, neither the denaturation/renaturation process nor the amino acid replacements have a noticeable effect on the three-dimensional structure of the BRU protease or on the detailed conformation of the catalytic site, which is very similar to that of other aspartyl proteases.
- URA 359 CNRS, Département d'Immunologie, Institut Pasteur, Paris, France.
Organizational Affiliation: