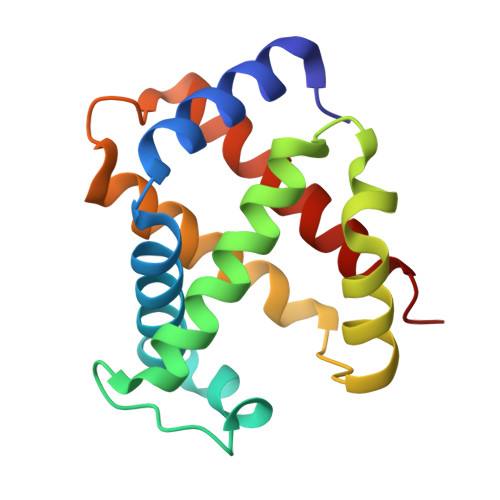
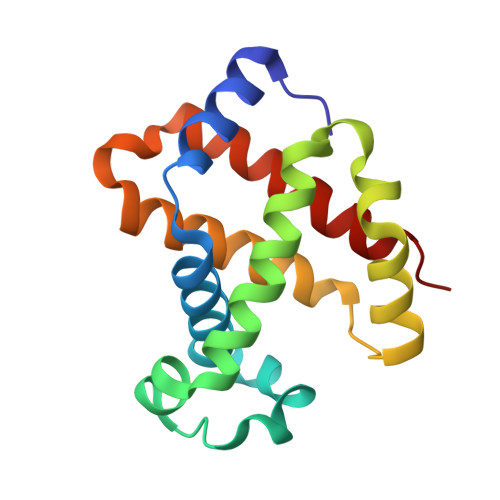
High resolution crystal structures and comparisons of T-state deoxyhaemoglobin and two liganded T-state haemoglobins: T(alpha-oxy)haemoglobin and T(met)haemoglobin.
Liddington, R., Derewenda, Z., Dodson, E., Hubbard, R., Dodson, G.(1992) J Mol Biology 228: 551-579
- PubMed: 1453464
- DOI: https://doi.org/10.1016/0022-2836(92)90842-8
- Primary Citation of Related Structures:
1HGA, 1HGB, 1HGC - PubMed Abstract:
The origin of co-operativity in haemoglobin (Hb) resides in the reduced affinity of the T-state. T-state Hb crystals grown from polyethyleneglycol can be liganded without the molecule switching to the R high affinity state. X-ray analysis of T-state alpha-oxy Hb and T-state met Hb has identified the structural basis for reduced affinity. The nature of the chemical tension at the haem environment is different in the alpha and beta haems. There are small but definite structural changes associated with ligation in the T-state: these prove to be mostly in the same direction as the larger changes that occur in the T-->R transition.
- Department of Chemistry, University of York, Heslington.
Organizational Affiliation: