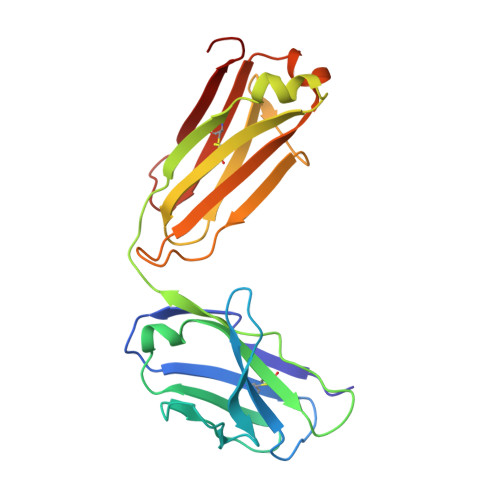
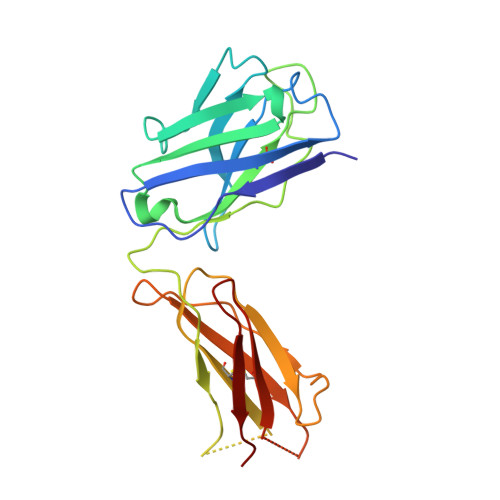
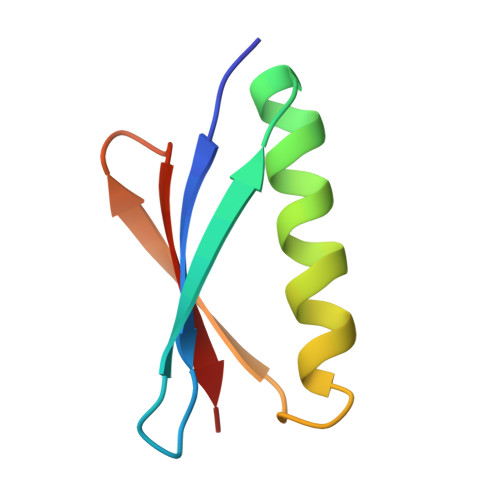
Complex between Peptostreptococcus Magnus Protein L and a Human Antibody Reveals Structural Convergence in the Interaction Modes of Fab Binding Modes
Graille, M., Stura, E.A., Housden, N.G., Beckingham, J.A., Bottomley, S.P., Beale, D., Taussig, M.J., Sutton, B.J., Gore, M.G., Charbonnier, J.B.(2001) Structure 9: 679
- PubMed: 11587642
- DOI: https://doi.org/10.1016/s0969-2126(01)00630-x
- Primary Citation of Related Structures:
1HEZ - PubMed Abstract:
Peptostreptococcus magnus protein L (PpL) is a multidomain, bacterial surface protein whose presence correlates with virulence. It consists of up to five homologous immunoglobulin binding domains that interact with the variable (VL) regions of kappa light chains found on two thirds of mammalian antibodies. We refined the crystal structure of the complex between a human antibody Fab fragment (2A2) and a single PpL domain (61 residues) to 2.7 A. The asymmetric unit contains two Fab molecules sandwiching a single PpL domain, which contacts similar VL framework regions of two light chains via independent interfaces. The residues contacted on VL are remote from the hypervariable loops. One PpL-Vkappa interface agrees with previous biochemical data, while the second is novel. Site-directed mutagenesis and analytical-centrifugation studies suggest that the two PpL binding sites have markedly different affinities for VL. The PpL residues in both interactions are well conserved among different Peptostreptococcus magnus strains. The Fab contact positions identified in the complex explain the high specificity of PpL for antibodies with kappa rather than lambda chains. The PpL-Fab complex shows the first interaction of a bacterial virulence factor with a Fab light chain outside the conventional combining site. Structural comparison with two other bacterial proteins interacting with the Fab heavy chain shows that PpL, structurally homologous to streptococcal SpG domains, shares with the latter a similar binding mode. These two bacterial surface proteins interact with their respective immunoglobulin regions through a similar beta zipper interaction.
- Département d'Ingénierie et d'Etudes des Protéines, Commissariat à l'Energie Atomique, Centre d'Etudes Saclay, F-91191, Gif-sur-Yvette, France.
Organizational Affiliation: