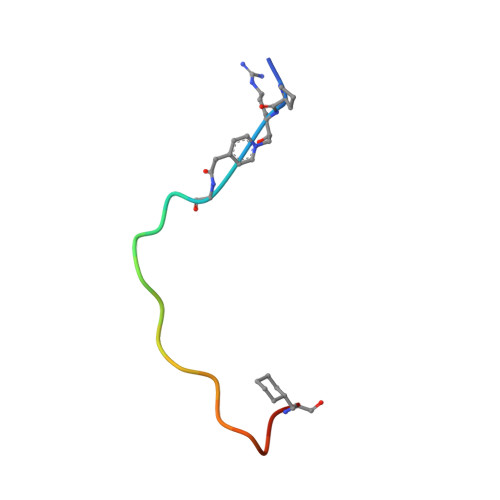
Crystal structure of a peptidyl pyridinium methyl ketone inhibitor with thrombin.
Rehse, P.H., Steinmetzer, T., Li, Y., Konishi, Y., Cygler, M.(1995) Biochemistry 34: 11537-11544
- PubMed: 7547884
- DOI: https://doi.org/10.1021/bi00036a029
- Primary Citation of Related Structures:
1HBT - PubMed Abstract:
The crystal structure of a complex between a bivalent peptidyl pyridinium methyl ketone inhibitor and human alpha-thrombin has been solved and refined at 2.0 A to an R factor of 0.18. The inhibitor, (D)cyclohexylalanine-Pro-Arg-(CH2N+C5H4CH2CO)-(Gly)4-Asp- Tyr-Glu-Pro-Ile-Pro-Glu-Glu-Ala-cyclo-hexylalanine-(D)Glu (coded P596), which forms a reversible covalent complex with thrombin, is highly potent with a Ki = 4.6 +/- 1.0 x 10(-14) M, lower than that of recombinant hirudin. The N-terminal, active-site-directed portion of the inhibitor is linked to the fibrinogen recognition exosite binding portion by a tetraglycine segment. The strong electron-withdrawing effect provided by the permanent positive charge on the pyridinium nitrogen makes the arginyl carbonyl carbon more susceptible to nucleophilic attack. In the crystal, a covalent P596-thrombin complex is observed. The electron density surrounding the active site portion and the pyridinium of the inhibitor is very well defined, clearly showing the existence of a covalent bond between the Ser195 O gamma and the now tetrahedral carbon of the inhibitor. The decreased binding ability of thrombin inhibitors containing N-terminal acetylation is discussed as is the effect of replacing the P3 (D)phenylalanine with (D)cyclohexylalanine. The electron density surrounding the remainder of the inhibitor is generally well defined, the exceptions being the C-terminal (D)Glu, the highly flexible tetraglycine linker, and some of the solvent-directed side chains.(ABSTRACT TRUNCATED AT 250 WORDS)
- Biotechnology Research Institute, National Research Council of Canada, Montreal, Quebec.
Organizational Affiliation: