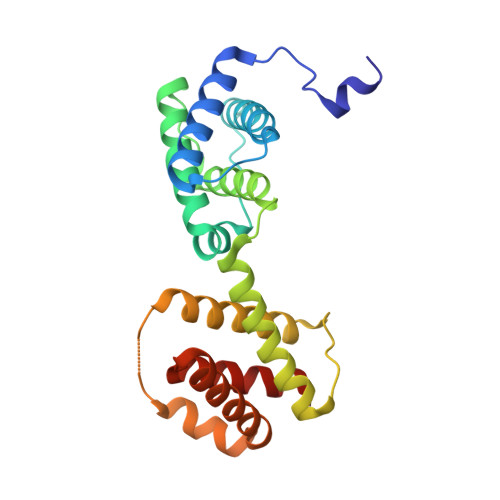
Crystal Structure of an Activated Form of the Pts Regulation Domain from the Lict Transcriptional Antitrminator
Van Tilbeurgh, H., Lecoq, D., Declerck, N.(2001) EMBO J 20: 3789
- PubMed: 11447120
- DOI: https://doi.org/10.1093/emboj/20.14.3789
- Primary Citation of Related Structures:
1H99 - PubMed Abstract:
The transcriptional antiterminator protein LicT regulates the expression of Bacillus subtilis operons involved in beta-glucoside metabolism. It belongs to a newly characterized family of bacterial regulators whose activity is controlled by the phosphoenolpyruvate:sugar phosphotransferase system (PTS). LicT contains an N-terminal RNA-binding domain (56 residues), and a PTS regulation domain (PRD, 221 residues) that is phosphorylated on conserved histidines in response to substrate availability. Replacement of both His207 and His269 with a negatively charged residue (aspartic acid) led to a highly active LicT variant that no longer responds to either induction or catabolite repression signals from the PTS. In contrast to wild type, the activated mutant form of the LicT regulatory domain crystallized easily and provided the first structure of a PRD, determined at 1.55 A resolution. The structure is a homodimer, each monomer containing two analogous alpha-helical domains. The phosphorylation sites are totally buried at the dimer interface and hence inaccessible to phosphorylating partners. The structure suggests important tertiary and quaternary rearrangements upon LicT activation, which could be communicated from the protein C-terminal end up to the RNA-binding domain.
- Architecture et Fonction des Macromolécules Biologiques, UMR 6098 du CNRS, Université d'Aix-Marseille, I et II, ESIL-GBMA, 163 Avenue de Luminy Case 925, 13288 Marseille Cedex 9, France. vantil@esil.univ-mrs.fr
Organizational Affiliation: