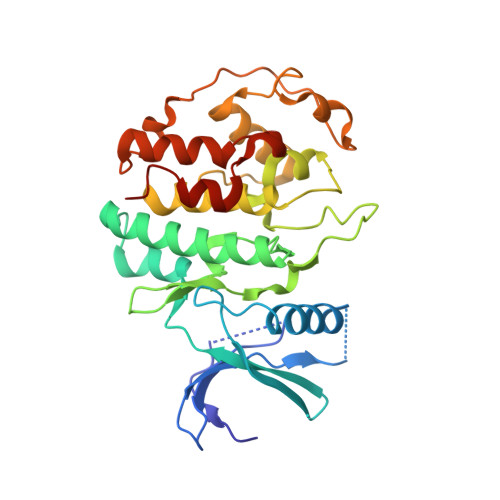
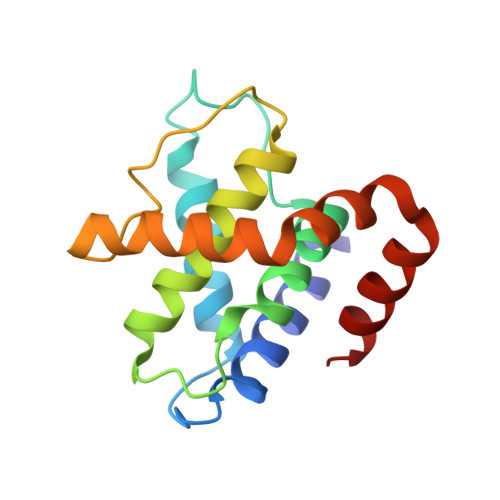
Structure and Regulation of the Cdk5-P25(Nck5A) Complex
Tarricone, C., Dhavan, R., Peng, J., Areces, L.B., Tsai, L.-H., Musacchio, A.(2001) Mol Cell 8: 657
- PubMed: 11583627
- DOI: https://doi.org/10.1016/s1097-2765(01)00343-4
- Primary Citation of Related Structures:
1H4L - PubMed Abstract:
CDK5 plays an indispensable role in the central nervous system, and its deregulation is involved in neurodegeneration. We report the crystal structure of a complex between CDK5 and p25, a fragment of the p35 activator. Despite its partial structural similarity with the cyclins, p25 displays an unprecedented mechanism for the regulation of a cyclin-dependent kinase. p25 tethers the unphosphorylated T loop of CDK5 in the active conformation. Residue Ser159, equivalent to Thr160 on CDK2, contributes to the specificity of the CDK5-p35 interaction. Its substitution with threonine prevents p35 binding, while the presence of alanine affects neither binding nor kinase activity. Finally, we provide evidence that the CDK5-p25 complex employs a distinct mechanism from the phospho-CDK2-cyclin A complex to establish substrate specificity.
- Structural Biology Unit, Department of Experimental Oncology, European Institute of Oncology, Via Ripamonti 435, I-20141 Milan, Italy.
Organizational Affiliation: