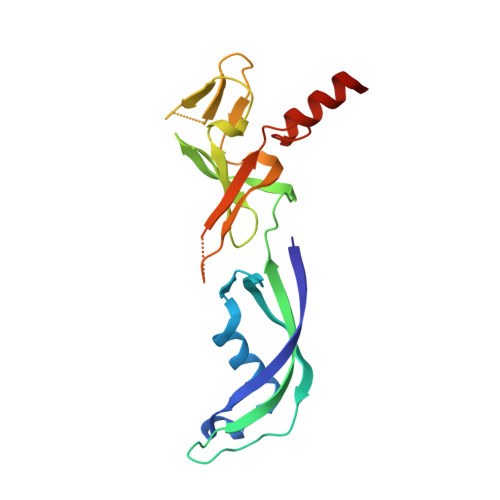
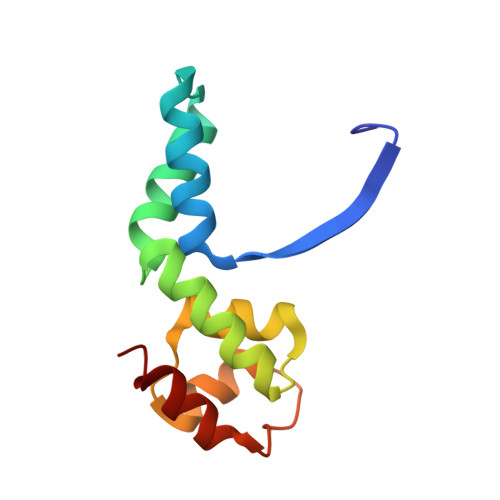
Structure of an Archaeal Homolog of the Eukaryotic RNA Polymerase II Rpb4/Rpb7 Complex
Todone, F., Brick, P., Werner, F., Weinzierl, R.O.J., Onesti, S.(2001) Mol Cell 8: 1137-1143
- PubMed: 11741548
- DOI: https://doi.org/10.1016/s1097-2765(01)00379-3
- Primary Citation of Related Structures:
1GO3 - PubMed Abstract:
The eukaryotic subunits RPB4 and RPB7 form a heterodimer that reversibly associates with the RNA polymerase II core and constitute the only two components of the enzyme for which no structural information is available. We have determined the crystal structure of the complex between the Methanococcus jannaschii subunits E and F, the archaeal homologs of RPB7 and RPB4. Subunit E has an elongated two-domain structure and contains two potential RNA binding motifs, while the smaller F subunit wraps around one side of subunit E, at the interface between the two domains. We propose a model for the interaction between RPB4/RPB7 and the core RNA polymerase in which the RNA binding face of RPB7 is positioned to interact with the nascent RNA transcript.
- Department of Biological Sciences, Imperial College, Exhibition Road, SW7 2AZ, London, United Kingdom.
Organizational Affiliation: