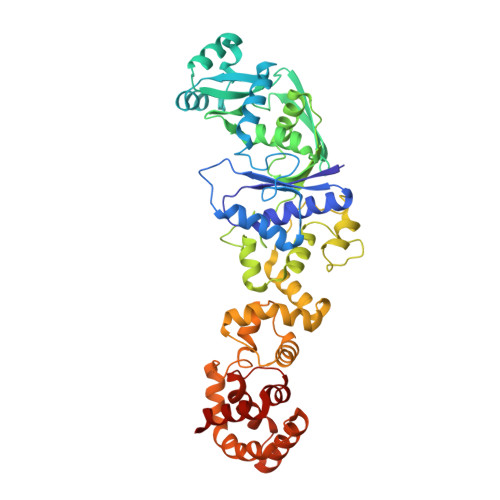
Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
Nureki, O., Vassylyev, D.G., Katayanagi, K., Shimizu, T., Sekine, S., Kigawa, T., Miyazawa, T., Yokoyama, S., Morikawa, K.(1995) Science 267: 1958-1965
- PubMed: 7701318
- DOI: https://doi.org/10.1126/science.7701318
- Primary Citation of Related Structures:
1GLN - PubMed Abstract:
The crystal structure of a class I aminoacyl-transfer RNA synthetase, glutamyl-tRNA synthetase (GluRS) from Thermus thermophilus, was solved and refined at 2.5 A resolution. The amino-terminal half of GluRS shows a geometrical similarity with that of Escherichia coli glutaminyl-tRNA synthetase (GlnRS) of the same subclass in class I, comprising the class I-specific Rossmann fold domain and the intervening subclass-specific alpha/beta domain. These domains were found to have two GluRS-specific, secondary-structure insertions, which then participated in the specific recognition of the D and acceptor stems of tRNA(Glu) as indicated by mutagenesis analyses based on the docking properties of GluRS and tRNA. In striking contrast to the beta-barrel structure of the GlnRS carboxyl-terminal half, the GluRS carboxyl-terminal half displayed an all-alpha-helix architecture, an alpha-helix cage, and mutagenesis analyses indicated that it had a role in the anticodon recognition.
- Department of Biophysics and Biochemistry, School of Science, University of Tokyo, Japan.
Organizational Affiliation: