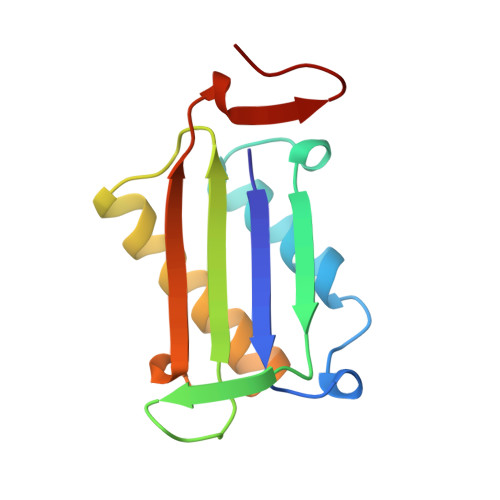
The crystal structure of human glycosylation-inhibiting factor is a trimeric barrel with three 6-stranded beta-sheets.
Kato, Y., Muto, T., Tomura, T., Tsumura, H., Watarai, H., Mikayama, T., Ishizaka, K., Kuroki, R.(1996) Proc Natl Acad Sci U S A 93: 3007-3010
- PubMed: 8610159
- DOI: https://doi.org/10.1073/pnas.93.7.3007
- Primary Citation of Related Structures:
1GIF - PubMed Abstract:
Glycosylation-inhibiting factor (GIF) is a cytokine that is involved in the regulation of IgE synthesis. The crystal structure of recombinant human GIF was determined by the multiple isomorphous replacement method. The structure was refined to an R factor of 0.168 at 1.9 angstrom resolution. The overall structure is seen to consist of three interconnected subunits forming a barrel with three 6-stranded beta-sheets on the inside and six alpha-helices on the outside. There is a 5-angstrom-diameter "hole" through the middle of the barrel. The barrel structure of GIF in part resembles other "trefoil" cytokines such as interleukin 1 and fibroblast growth factor. Each subunit has a new class of alpha + beta sandwich structure consisting of two beta-alpha-beta motifs. These beta-alpha-beta motifs are related by a pseudo-twofold axis and resemble both interleukin 8 and the peptide binding domain of major histocompatibility complex protein, although the topology of the polypeptide chain is quite different.
- Central Laboratories for Key Technology, Kirin Brewery Company, Ltd., Yokohama, Japan.
Organizational Affiliation: