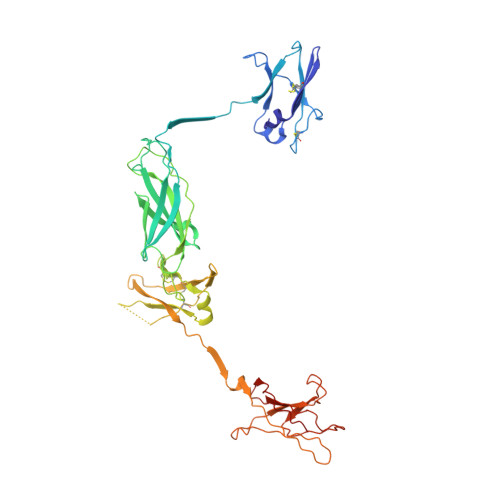
Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration.
Carr, P.D., Gustin, S.E., Church, A.P., Murphy, J.M., Ford, S.C., Mann, D.A., Woltring, D.M., Walker, I., Ollis, D.L., Young, I.G.(2001) Cell 104: 291-300
- PubMed: 11207369
- DOI: https://doi.org/10.1016/s0092-8674(01)00213-6
- Primary Citation of Related Structures:
1GH7 - PubMed Abstract:
The receptor systems for the hemopoietic cytokines GM-CSF, IL-3, and IL-5 consist of ligand-specific alpha receptor subunits that play an essential role in the activation of the shared betac subunit, the major signaling entity. Here, we report the structure of the complete betac extracellular domain. It has a structure unlike any class I cytokine receptor described thus far, forming a stable interlocking dimer in the absence of ligand in which the G strand of domain 1 hydrogen bonds into the corresponding beta sheet of domain 3 of the dimer-related molecule. The G strand of domain 3 similarly partners with the dimer-related domain 1. The structure provides new insights into receptor activation by the respective alpha receptor:ligand complexes.
- Research School of Chemistry, Australian National University, Acton, ACT 0200, Australia.
Organizational Affiliation: