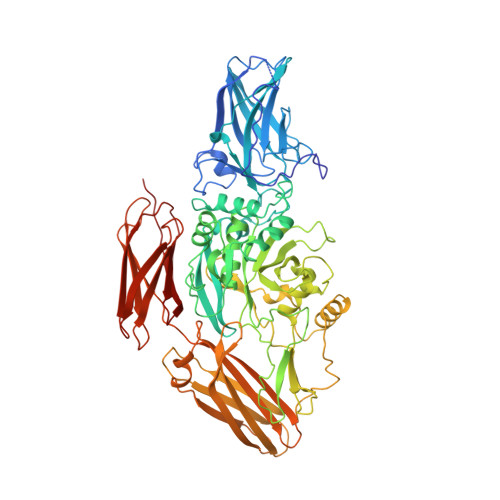
Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII.
Yee, V.C., Pedersen, L.C., Le Trong, I., Bishop, P.D., Stenkamp, R.E., Teller, D.C.(1994) Proc Natl Acad Sci U S A 91: 7296-7300
- PubMed: 7913750
- DOI: https://doi.org/10.1073/pnas.91.15.7296
- Primary Citation of Related Structures:
1GGT - PubMed Abstract:
Mechanical stability in many biological materials is provided by the crosslinking of large structural proteins with gamma-glutamyl-epsilon-lysyl amide bonds. The three-dimensional structure of human recombinant factor XIII (EC 2.3.2.13 zymogen; protein-glutamine:amine gamma-glutamyltransferase a chain), a transglutaminase zymogen, has been solved at 2.8-A resolution by x-ray crystallography. This structure shows that each chain of the homodimeric protein is folded into four sequential domains. A catalytic triad reminiscent of that observed in cysteine proteases has been identified in the core domain. The amino-terminal activation peptide of each subunit crosses the dimer interface and partially occludes the opening of the catalytic cavity in the second subunit, preventing substrate binding to the zymogen. A proposal for the mechanism of activation by thrombin and calcium is made that details the structural events leading to active factor XIIIa'.
- Department of Biochemistry, University of Washington, Seattle 98195.
Organizational Affiliation: