Crystallization and preliminary X-ray analysis of a DNA dodecamer containing 2'-deoxy-5-formyluridine; what is the role of magnesium cation in crystallization of Dickerson-type DNA dodecamers?
Tsunoda, M., Karino, N., Ueno, Y., Matsuda, A., Takenaka, A.(2001) Acta Crystallogr D Biol Crystallogr 57: 345-348
- PubMed: 11173500
- DOI: https://doi.org/10.1107/s0907444900017583
- Primary Citation of Related Structures:
1G75, 1G8N, 1G8U, 1G8V - PubMed Abstract:
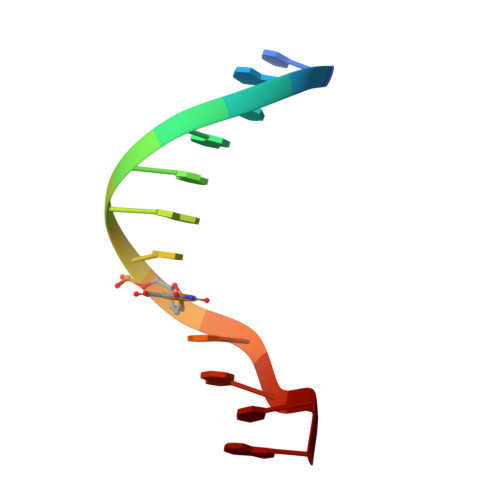
To investigate the role of divalent cations in crystal packing, four different crystals of a Dickerson-type dodecamer with the sequence d(CGCGAATXCGCG), containing 2'-deoxy-5-formyluridine at X, were obtained under several conditions with and without divalent cations. The crystal structures are all isomorphous. The octahedrally hydrated magnesium cations found in the major groove cement the two neighbouring duplexes along the b axis. In the Mg(2+)-free crystals, a five-membered ring of water molecules occupies the same position as the magnesium site and connects the two duplexes similarly to the hydrated Mg(2+) ion. It has been concluded that water molecules can take the place of the hydrated magnesium cation in crystallization, but the magnesium cation is more effective and gives X-ray diffraction at slightly higher resolution. In all four crystals, the 5-formyluracil residues form the canonical Watson-Crick pair with adenine residues.
- Department of Life Science, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Midori-ku, Yokohama 226-8501, Japan.
Organizational Affiliation: