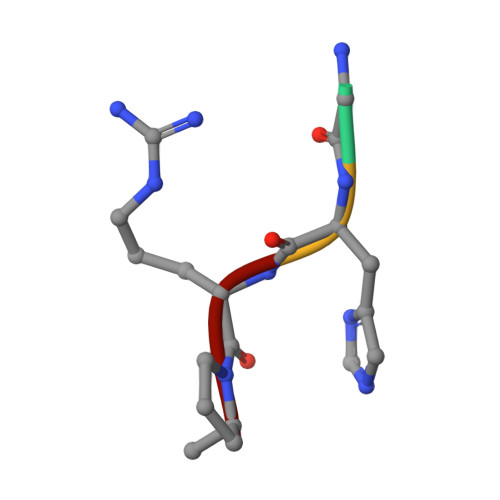
Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Everse, S.J., Spraggon, G., Veerapandian, L., Doolittle, R.F.(1999) Biochemistry 38: 2941-2946
- PubMed: 10074346
- DOI: https://doi.org/10.1021/bi982626w
- Primary Citation of Related Structures:
1FZE, 1FZF, 1FZG - PubMed Abstract:
The structure of fragment double-D from human fibrin has been solved in the presence and absence of the peptide ligands that simulate the two knobs exposed by the removal of fibrinopeptides A and B, respectively. All told, six crystal structures have been determined, three of which are reported here for the first time: namely, fragments D and double-D with the peptide GHRPam alone and double-D in the absence of any peptide ligand. Comparison of the structures has revealed a series of conformational changes that are brought about by the various knob-hole interactions. Of greatest interest is a moveable "flap" of two negatively charged amino acids (Glubeta397 and Aspbeta398) whose side chains are pinned back to the coiled coil with a calcium atom bridge until GHRPam occupies the beta-chain pocket. Additionally, in the absence of the peptide ligand GPRPam, GHRPam binds to the gamma-chain pocket, a new calcium-binding site being formed concomitantly.
- Center for Molecular Genetics, University of California, San Diego, La Jolla 92093-0634, USA.
Organizational Affiliation: