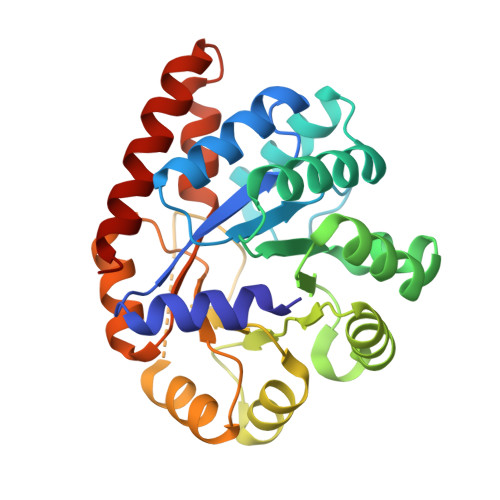
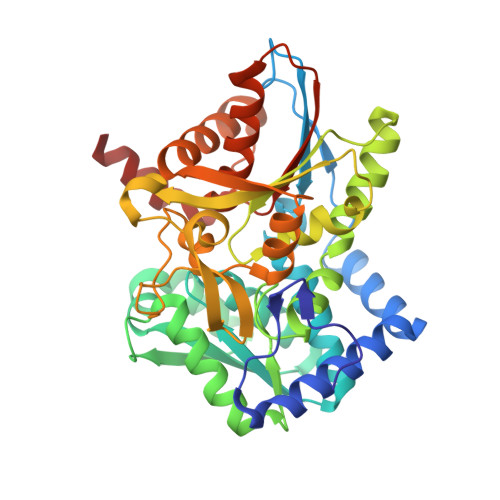
Structural basis for the impaired channeling and allosteric inter-subunit communication in the beta A169L/beta C170W mutant of tryptophan synthase.
Weyand, M., Schlichting, I.(2000) J Biological Chem 275: 41058-41063
- PubMed: 11034989
- DOI: https://doi.org/10.1074/jbc.C000479200
- Primary Citation of Related Structures:
1FUY - PubMed Abstract:
We determined the 2.25 A resolution crystal structure of the betaA169L/betaC170W mutant form of the tryptophan synthase alpha(2)beta(2) complex from Salmonella typhimurium complexed with the alpha-active site substrate analogue 5-fluoro-indole-propanol-phosphate to identify the structural basis for the changed kinetic properties of the mutant (Anderson, K. S., Kim, A. Y., Quillen, J. M., Sayers, E., Yang, X. J., and Miles, E. W. (1995) J. Biol. Chem. 270, 29936-29944). Comparison with the wild-type enzyme showed that the betaTrp(170) side chain occludes the tunnel connecting the alpha- and beta-active sites, explaining the accumulation of the intermediate indole during a single enzyme turnover. To prevent a steric clash between betaLeu(169) and betaGly(135), located in the beta-sheet of the COMM (communication) domain (betaGly(102)-betaGly(189)), the latter reorganizes. The changed COMM domain conformation results in a loss of the hydrogen bonding networks between the alpha- and beta-active sites, explaining the poor activation of the alpha-reaction upon formation of the aminoacrylate complex at the beta-active site. The 100-fold reduced affinity for serine seems to result from a movement of betaAsp(305) away from the beta-active site so that it cannot interact with the hydroxyl group of a pyridoxal phosphate-bound serine. The proposed structural dissection of the effects of each single mutation in the betaA169L/betaC170W mutant would explain the very different kinetics of this mutant and betaC170F.
- Max Planck Institut für Molekulare Physiologie, Abteilung Physikalische Biochemie, Otto-Hahn-Strabetae 11, 44227 Dortmund, Germany.
Organizational Affiliation: