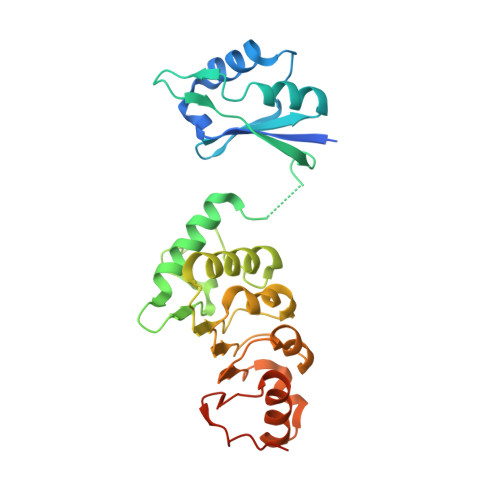
The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain.
Liker, E., Fernandez, E., Izaurralde, E., Conti, E.(2000) EMBO J 19: 5587-5598
- PubMed: 11060011
- DOI: https://doi.org/10.1093/emboj/19.21.5587
- Primary Citation of Related Structures:
1FO1, 1FT8 - PubMed Abstract:
Human TAP is implicated in mRNA nuclear export and is used by simian type D retroviruses to export their unspliced genomic RNA to the cytoplasm of the host cell. We have determined the crystal structure of the minimal TAP fragment that binds the constitutive transport element (CTE) of retroviral RNAs. Unexpectedly, we find the fragment consists of a ribonucleoprotein (RNP) domain, which is not identifiable by its sequence, and a leucine-rich repeat (LRR) domain. The non-canonical RNP domain functions as the general RNA-binding portion of the fragment. The LRR domain is required in cis to the RNP domain for CTE RNA binding. The structural and biochemical properties of the domains point to a remarkable similarity with the U2B"(RNP)-U2A'(LRR) spliceosomal heterodimer. Our in vitro and in vivo functional studies using structure-based mutants suggest that a phylogenetically conserved surface of the LRR domain of TAP may have different roles in the export of viral and cellular RNAs.
- European Molecular Biology Laboratory (EMBL), Meyerhofstrasse 1, D-69117 Heidelberg, Germany. conti@embl-heidelberg.de
Organizational Affiliation: