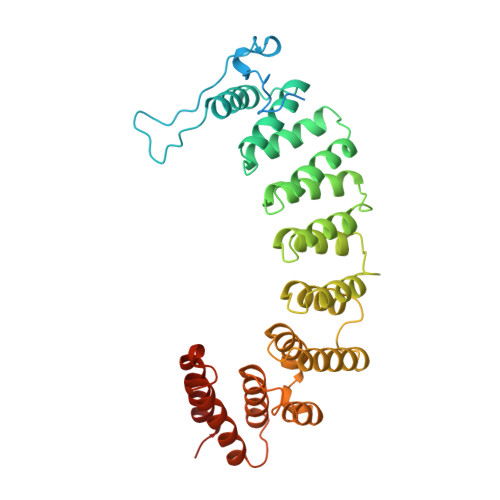
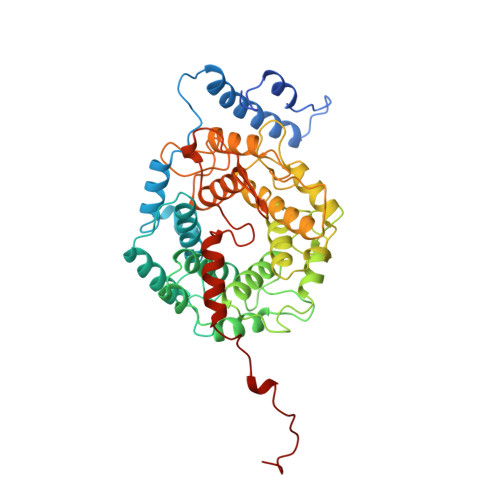
Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution.
Park, H.W., Boduluri, S.R., Moomaw, J.F., Casey, P.J., Beese, L.S.(1997) Science 275: 1800-1804
- PubMed: 9065406
- DOI: https://doi.org/10.1126/science.275.5307.1800
- Primary Citation of Related Structures:
1FT1 - PubMed Abstract:
Protein farnesyltransferase (FTase) catalyzes the carboxyl-terminal lipidation of Ras and several other cellular signal transduction proteins. The essential nature of this modification for proper function of these proteins has led to the emergence of FTase as a target for the development of new anticancer therapy. Inhibition of this enzyme suppresses the transformed phenotype in cultured cells and causes tumor regression in animal models. The crystal structure of heterodimeric mammalian FTase was determined at 2.25 angstrom resolution. The structure shows a combination of two unusual domains: a crescent-shaped seven-helical hairpin domain and an alpha-alpha barrel domain. The active site is formed by two clefts that intersect at a bound zinc ion. One cleft contains a nine-residue peptide that may mimic the binding of the Ras substrate; the other cleft is lined with highly conserved aromatic residues appropriate for binding the farnesyl isoprenoid with required specificity.
- Department of Biochemistry, Duke University Medical Center, Durham, NC 27710, USA.
Organizational Affiliation: