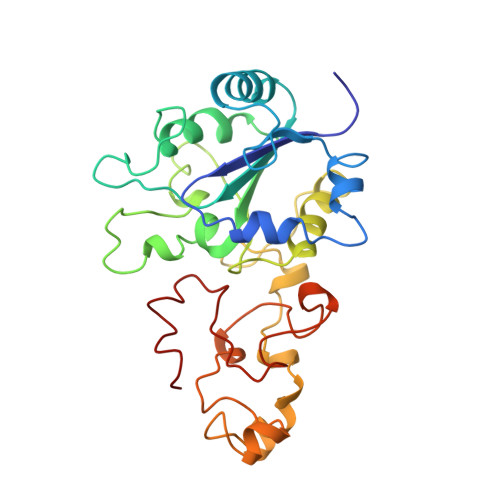
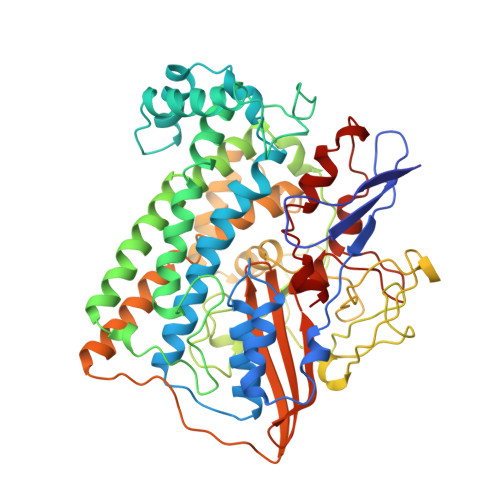
Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas.
Volbeda, A., Charon, M.H., Piras, C., Hatchikian, E.C., Frey, M., Fontecilla-Camps, J.C.(1995) Nature 373: 580-587
- PubMed: 7854413
- DOI: https://doi.org/10.1038/373580a0
- Primary Citation of Related Structures:
1FRV - PubMed Abstract:
The X-ray structure of the heterodimeric Ni-Fe hydrogenase from Desulfovibrio gigas, the enzyme responsible for the metabolism of molecular hydrogen, has been solved at 2.85 A resolution. The active site, which appears to contain, besides nickel, a second metal ion, is buried in the 60K subunit. The 28K subunit, which coordinates one [3Fe-4S] and two [4Fe-4S] clusters, contains an amino-terminal domain with similarities to the redox protein flavodoxin. The structure suggests plausible electron and proton transfer pathways.
- Laboratoire de Cristallographie et de Cristallogénèse des Protéines, Institut de Biologie Structurale J. P. Ebel (CEA, CNRS), Grenoble, France.
Organizational Affiliation: