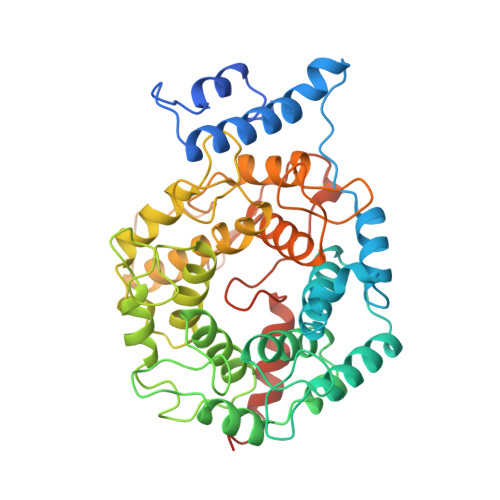
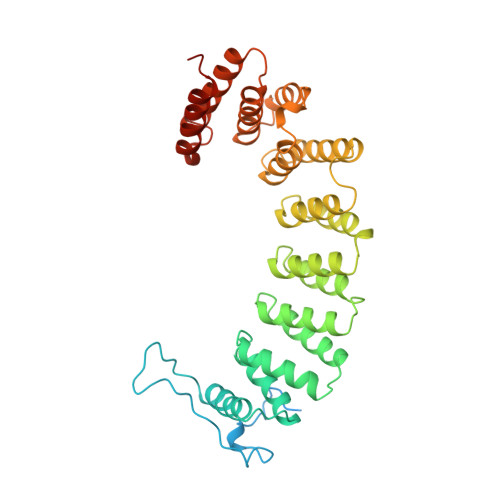
Protein farnesyltransferase: structure and implications for substrate binding.
Dunten, P., Kammlott, U., Crowther, R., Weber, D., Palermo, R., Birktoft, J.(1998) Biochemistry 37: 7907-7912
- PubMed: 9609683
- DOI: https://doi.org/10.1021/bi980531o
- Primary Citation of Related Structures:
1FPP - PubMed Abstract:
The rat protein farnesyltransferase crystal structure has been solved by multiple isomorphous replacement methods at a resolution of 2.75 A. The three-dimensional structure, together with recent data on the effects of several mutations, led us to propose a model for substrate binding which differs from the model presented by Park et al. based on their independent structure determination [Park, H. -W., Boduluri, S. R., Moomaw, J. F., Casey, P. J., and Beese, L. S. (1997) Science 275, 1800-1804]. Both farnesyl diphosphate and peptide substrates can be accommodated in the hydrophobic active-site barrel, with the sole charged residue inside the barrel, Arg202 of the beta-subunit, forming a salt bridge with the negatively charged carboxy terminus of peptide substrates. Our proposals are based in part on the observation of electron density in the active site which can be modeled as bound farnesyl diphosphate carried through the enzyme purification. In addition, our model explains in structural terms the results of mutational studies which have identified several residues critical for substrate specificity and catalysis.
- Roche Research Center, Hoffmann-La Roche Inc., Nutley, New Jersey 07110, USA.
Organizational Affiliation: