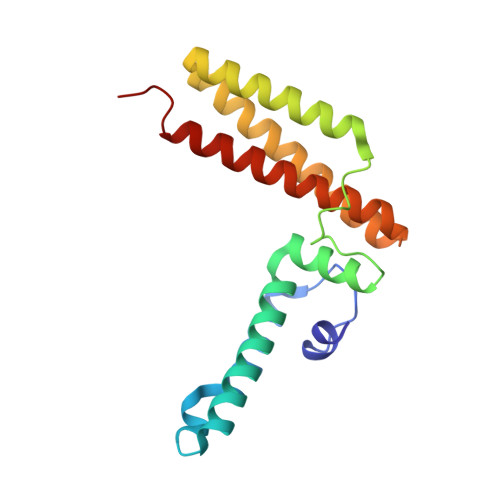
Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli.
Cupp-Vickery, J.R., Vickery, L.E.(2000) J Mol Biology 304: 835-845
- PubMed: 11124030
- DOI: https://doi.org/10.1006/jmbi.2000.4252
- Primary Citation of Related Structures:
1FPO - PubMed Abstract:
Hsc20 is a 20 kDa J-protein that regulates the ATPase activity and peptide-binding specificity of Hsc66, an hsp70-class molecular chaperone. We report herein the crystal structure of Hsc20 from Escherichia coli determined to a resolution of 1.8 A using a combination of single isomorphous replacement (SIR) and multi-wavelength anomalous diffraction (MAD). The overall structure of Hsc20 consists of two distinct domains, an N-terminal J-domain containing residues 1-75 connected by a short loop to a C-terminal domain containing residues 84-171. The structure of the J-domain, involved in interactions with Hsc66, resembles the alpha-topology of J-domain fragments of Escherichia coli DnaJ and human Hdj1 previously determined by solution NMR methods. The C-terminal domain, implicated in binding and targeting proteins to Hsc66, consists of a three-helix bundle in which two helices comprise an anti-parallel coiled-coil. The two domains make contact through an extensive hydrophobic interface ( approximately 650 A(2)) suggesting that their relative orientations are fixed. Thus, Hsc20, in addition to its role in the regulation of the ATPase activity of Hsc66, may also function as a rigid scaffold to facilitate positioning of the protein substrates targeted to Hsc66.
- Department of Chemistry and Biochemistry, California State University, Fullerton, CA 92834, USA. lvickery@uci.edu
Organizational Affiliation: