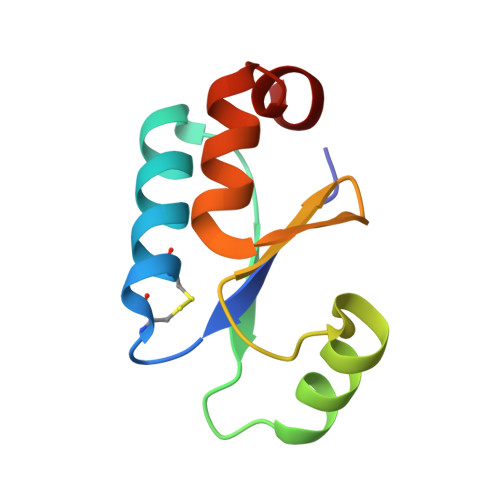
NMR structure of oxidized glutaredoxin 3 from Escherichia coli.
Nordstrand, K., Sandstrom, A., Aslund, F., Holmgren, A., Otting, G., Berndt, K.D.(2000) J Mol Biology 303: 423-432
- PubMed: 11031118
- DOI: https://doi.org/10.1006/jmbi.2000.4145
- Primary Citation of Related Structures:
1FOV - PubMed Abstract:
A high precision NMR structure of oxidized glutaredoxin 3 [C65Y] from Escherichia coli has been determined. The conformation of the active site including the disulphide bridge is highly similar to those in glutaredoxins from pig liver and T4 phage. A comparison with the previously determined structure of glutaredoxin 3 [C14S, C65Y] in a complex with glutathione reveals conformational changes between the free and substrate-bound form which includes the sidechain of the conserved, active site tyrosine residue. In the oxidized form this tyrosine is solvent exposed, while it adopts a less exposed conformation, stabilized by hydrogen bonds, in the mixed disulfide with glutathione. The structures further suggest that the formation of a covalent linkage between glutathione and glutaredoxin 3 is necessary in order to induce these structural changes upon binding of the glutathione peptide. This could explain the observed low affinity of glutaredoxins for S-blocked glutathione analogues, in spite of the fact that glutaredoxins are highly specific reductants of glutathione mixed disulfides.
- Center for Structural Biochemistry Karolinska Institutet, Huddinge, S-141 57, Sweden.
Organizational Affiliation: